Atp7b Gene Large Fragment Deletion Detection Kit and Detection Method
A technology for detection kits and large fragments, which is applied in the field of ATP7B gene large fragment deletion detection kits, can solve the problems of long import time, high price, and restrictions on wide application, so as to reduce the use threshold and cost, ensure efficiency and accuracy The effect of the detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment l
[0050] Probe and Primer Design
[0051] The present invention designs relevant probes for the 5'UTR promoter region and 21 exons of the ATP7B gene, and each site includes a pair of probes (left probe and right probe). The left probe sequentially (5'-3') includes an upstream universal primer binding sequence, a stuffer sequence of varying lengths, and a target-specific sequence; the right probe sequentially (5'-3') includes a target-specific sequence , stuffer sequences of varying lengths and downstream universal primer binding sequences. The probe sequences are shown in Table 1.
[0052] Table 1 ATP7B gene large fragment deletion detection probe sequence
[0053]
[0054]
[0055]
[0056]
[0057] The left and right probes are prepared by chemical synthesis, and the right probe is modified with a phosphorylation group at the 5' end during synthesis, so as to complete the connection reaction with the free hydroxyl group at the 3' end of the left probe. The synth...
Embodiment 2
[0063] Detection of ATP7B Gene Large Fragment Deletion Mutations in Healthy Human Samples
[0064] (1) Sample DNA extraction
[0065] To obtain peripheral blood DNA, use a blood genomic DNA extraction kit (such as Tiangen Biochemical Technology No. DP348 product), and follow the kit instructions to extract genomic DNA. Detect the concentration and purity of DNA, and adjust the DNA concentration to 20ng / μl.
[0066] (2) Genomic DNA denaturation
[0067] Each detection sample is divided into 4 tubes for detection, and each tube detects 6 sites. Take 5 μl of DNA sample (100 ng) from each reaction tube and incubate at 95°C for 5 minutes on a PCR machine, then rapidly cool to room temperature (25°C) to denature the genomic DNA into a single strand.
[0068] (3) Hybridization of probes to genomic DNA
[0069] Add 1.5 μl Taq high-fidelity DNA ligase (for example, product number M0647S from NEB Company) Buffer, 6 μl probe mixed solution (0.5 μl for each probe, 5 μM) and 2.5 μl DEP...
Embodiment 3
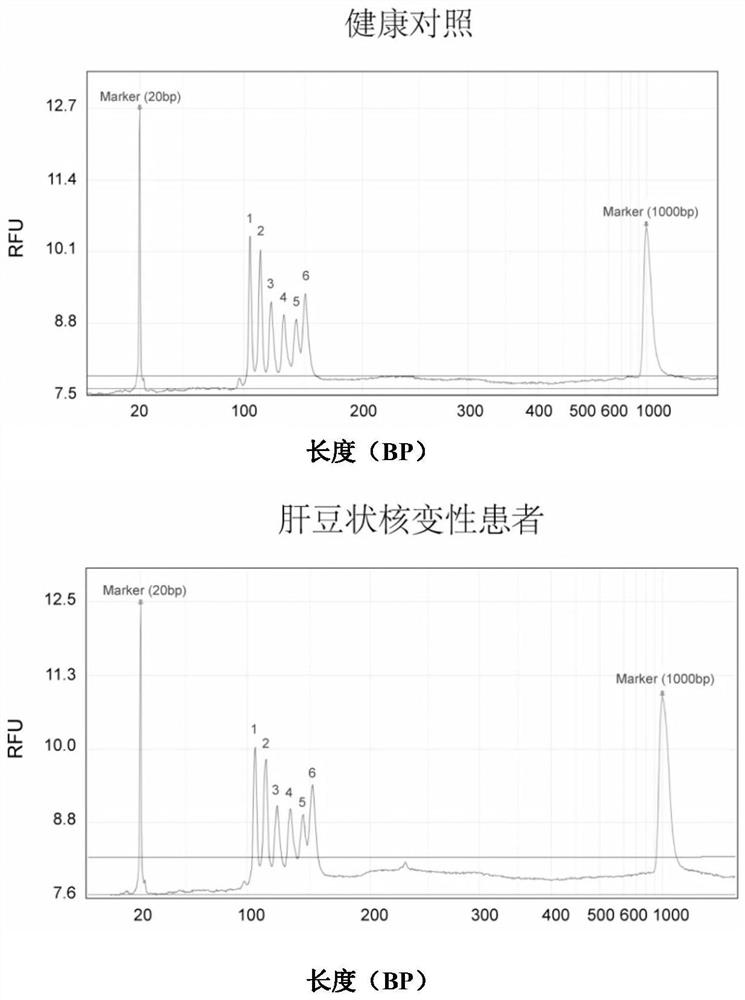
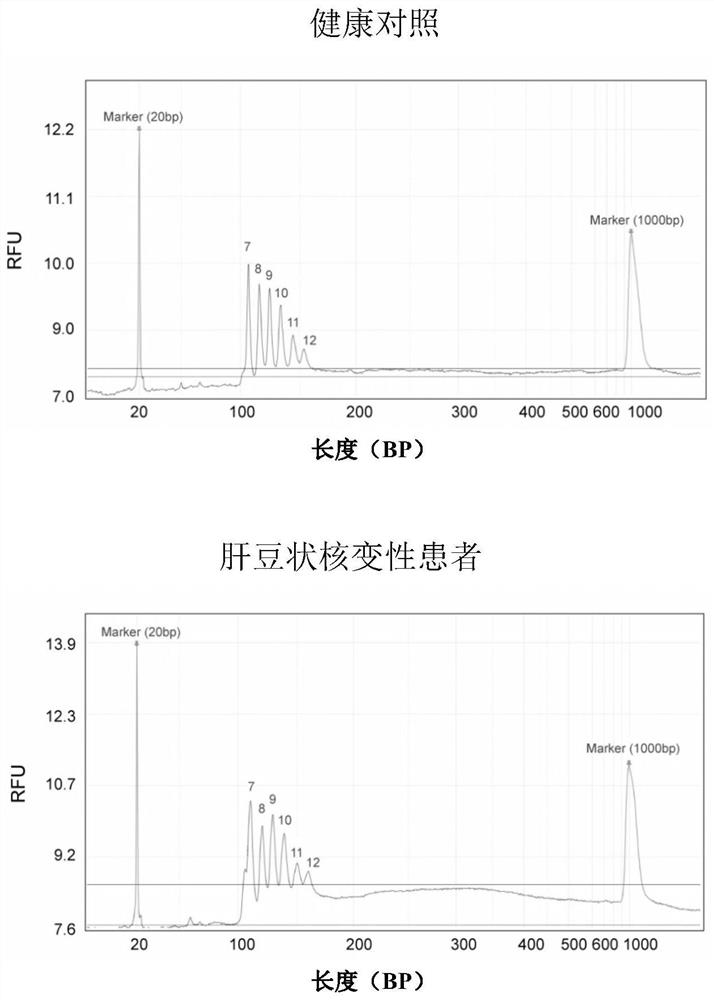
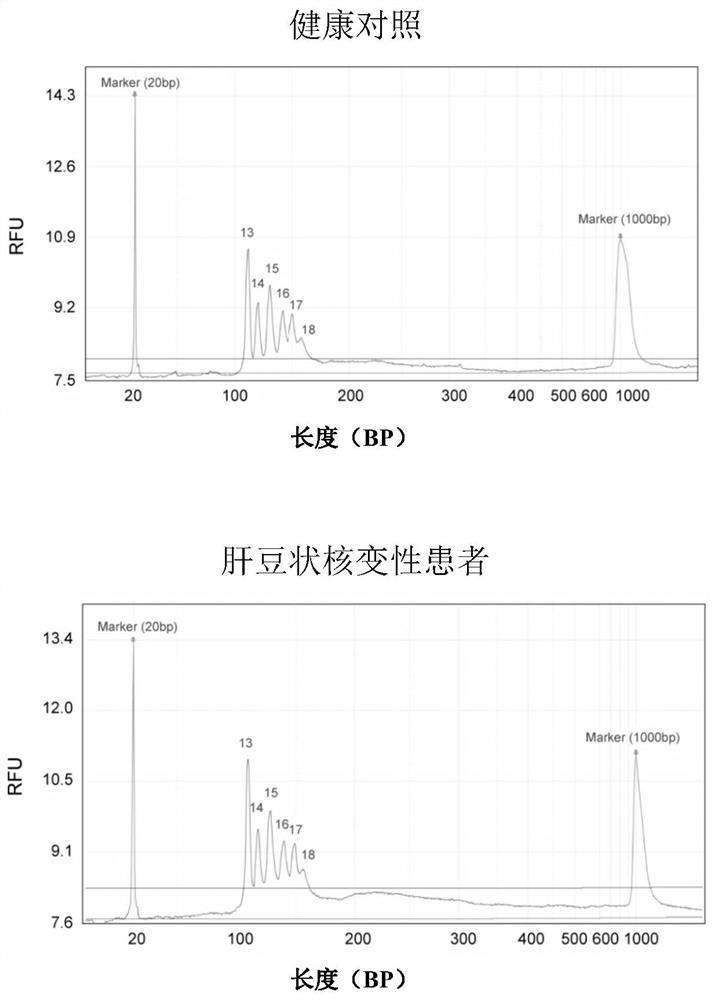
[0099] Detection of large fragment deletion mutations of ATP7B gene in patients with hepatolenticular degeneration
[0100] Steps (1)-(5) are the same as in Example 2.
[0101] (6) Detection of amplification products by capillary electrophoresis
[0102]Take the PCR product and detect it in the Qsep100 automatic nucleic acid analysis system, using a high-resolution sampling cartridge (S1 cartridge), the sampling voltage is 4KV, the sampling time is 10 seconds, the electrophoresis voltage is 6KV, and 20bp and 1000bp DNA markers are added at the same time . After the product concentration is converted into an electrical signal by the supporting software, the peak diagram is given, and the peak diagram ordinate RFU value is read, and compared with the RFU value of the healthy control in Example 2, if the RFU value is 50% of the control group or has no value, Then it can be judged that there is a heterozygous or homozygous deletion at the corresponding site of the ATP7B gene in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com