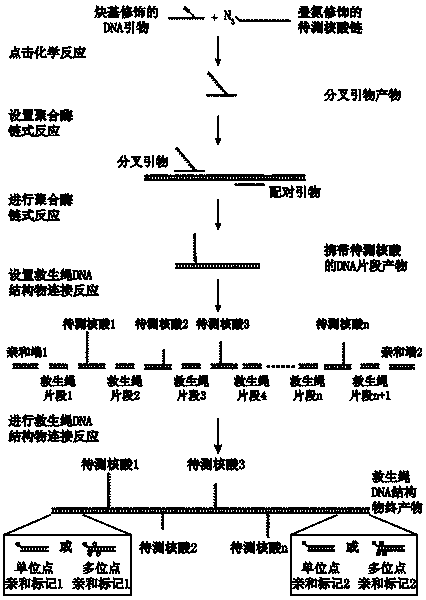
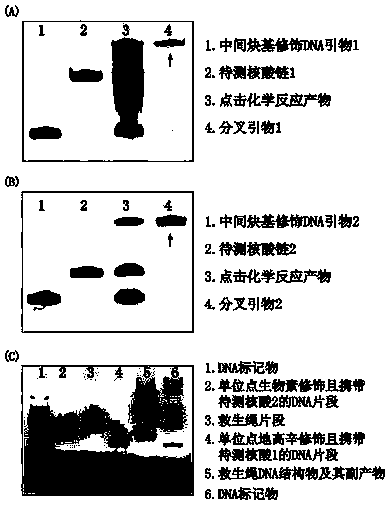
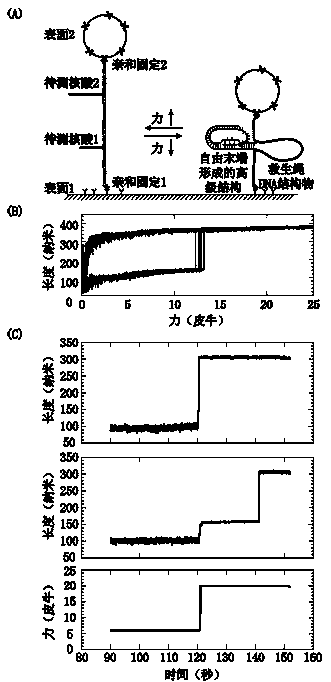
Method for detecting nucleic acid terminal structure based on single-molecule force spectroscopy
A technology of single-molecule force spectroscopy and nucleic acid, which is applied in the field of mechanical precision measurement of biomacromolecule secondary or higher three-dimensional conformation or advanced structure, and can solve problems that have not been reported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The present invention will be described in further detail below in conjunction with specific embodiments. For the experimental methods that do not indicate specific conditions in the examples, usually follow the conventional conditions and the conditions described in the manual, or according to the conditions suggested by the manufacturer; the general equipment, materials, reagents, etc. used can be used without special instructions. Obtained commercially.
[0028] The embodiment uses click chemistry, two single-stranded DNAs to be tested, single-site modification of biotin and digoxigenin, and single-molecule magnetic tweezers to describe the present invention in detail.
[0029] Table 1 Oligonucleotide name and sequence information
[0030] Oligonucleotide name
Sequence (5′ — 3′)
Remark
Nucleic acid strand to be tested 1
TTTTATCAGATTTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGATGTAGTATGTTGAGTGT
5' azide modification
Intermediate Alkynyl M...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com