Molecular markers for rice cold-tolerant major QTL qCTS12 and identification method and application of molecular markers
A technology of molecular markers and identification methods, applied in the field of molecular genetics, can solve the problems of limited residence and fine-mapping QTL, and achieve the effects of accelerating the breeding process, facilitating cross-breeding, and clear positions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Screening and mapping of molecular markers
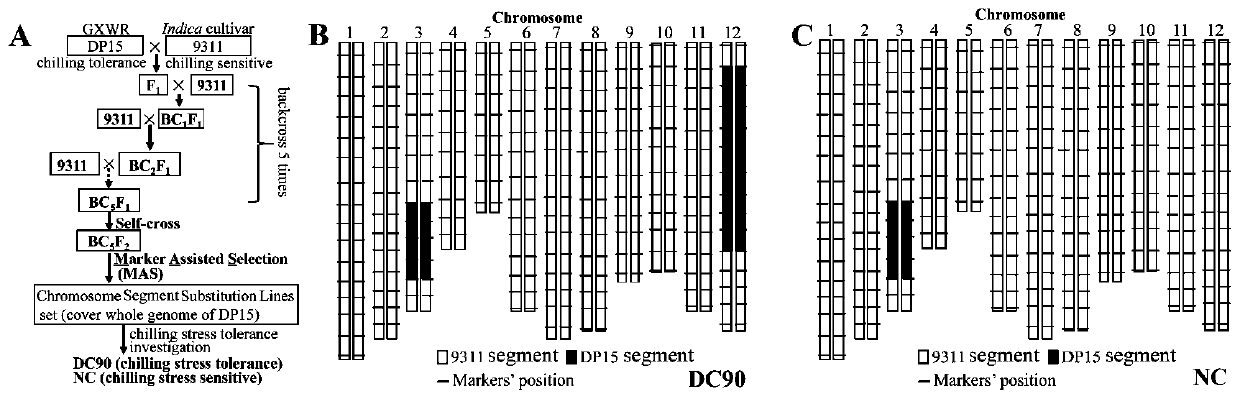
[0040] 1. Construction of the cold-tolerant chromosome segment substitution line CSSL12 and analysis of the cold-resistant phenotype of the replaced segment
[0041] (1) Using the cold-tolerant Guangxi common wild rice DP15 as the donor parent and the cold-sensitive rice variety 9311 as the recurrent parent to construct chromosome segment substitution lines (chromosome segment substitution lines, CSSLs), through continuous backcrossing, selfing and orientation Choice, in BC 5 f 2 Two chromosomal segment replacement lines with similar genetic backgrounds, DC90 and NC, were obtained. Most of the genetic background of DC90 is the same as that of the recurrent parent 9311, only the fragment replacement of DP15 on chromosome 3 and chromosome 12, the length of which is 10Mb and 20Mb respectively. However, NC only has a fragment replacement of DP15 on chromosome 3, which is the same as the replacement region of chromosome 3 in DC90...
Embodiment 2
[0063] Validation of Molecular Markers
[0064] 1. Materials and methods
[0065] 1.1 Materials
[0066] 52 rice varieties from different sources
[0067] 1.2 Method
[0068] 1.2.1 PCR amplification: Genomic DNA of rice leaves was extracted by the TPS extraction method, and the extracted sample DNA was amplified with primers M5 and M6, and the method was the same as in Example 1.
[0069] 1.2.2 Identification of cold stress phenotypes: select full-bodied seeds and germinate in water at 30°C for 3 days. After the seeds are "white", they are sown in plastic buckets filled with field soil to grow normally, the ambient temperature is 30°C, the light is 20000 Lux, and the air humidity is 40%. Cold treatment was carried out when the seedlings grew to the 3-leaf stage. When the seedlings grow to the 3-leaf stage, put them into an artificial climate culture room at 8°C-10°C for cold treatment for 5 days, during which the photoperiod is 13h (light) / 11h (dark), the light intensity ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com