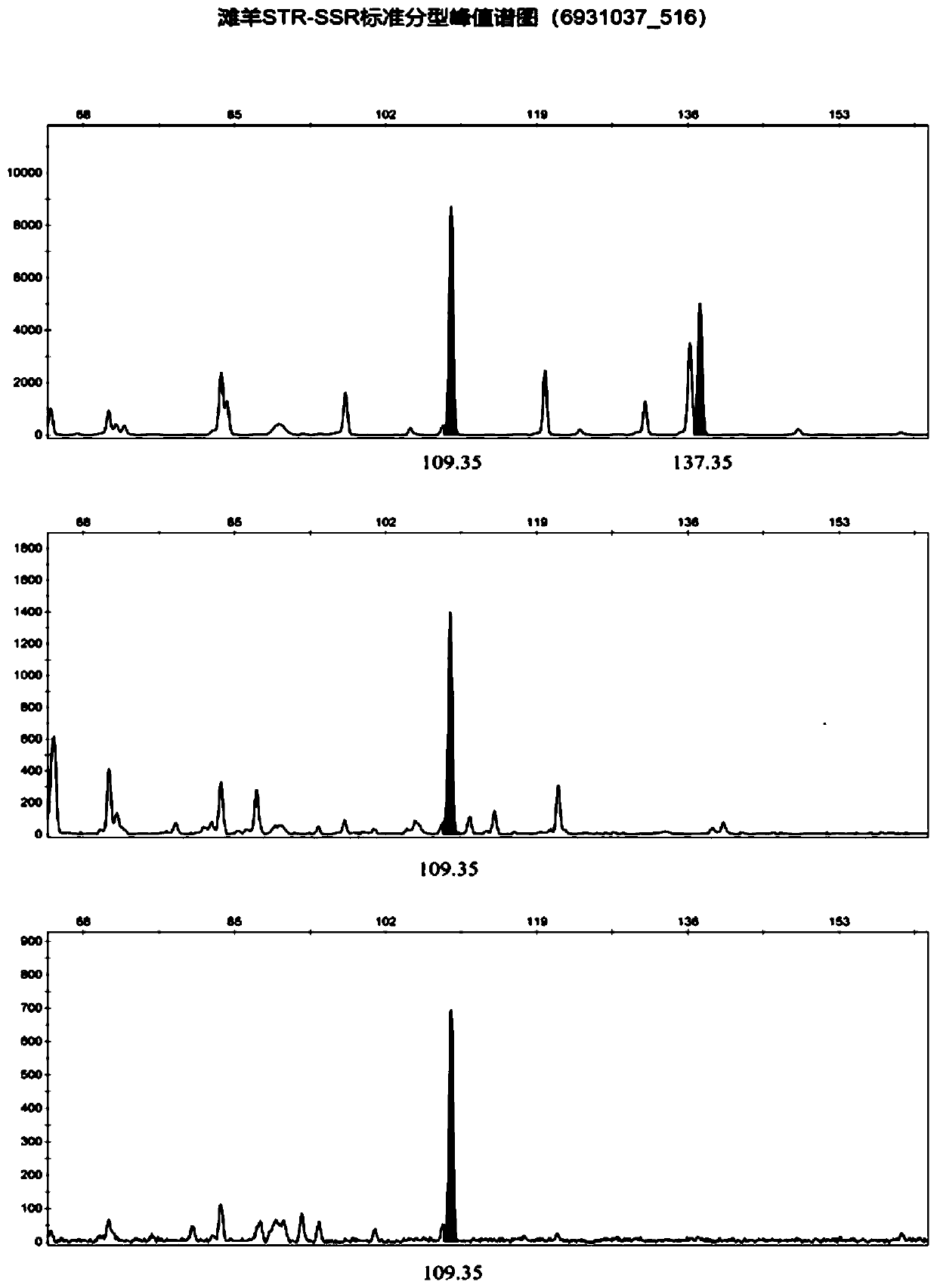
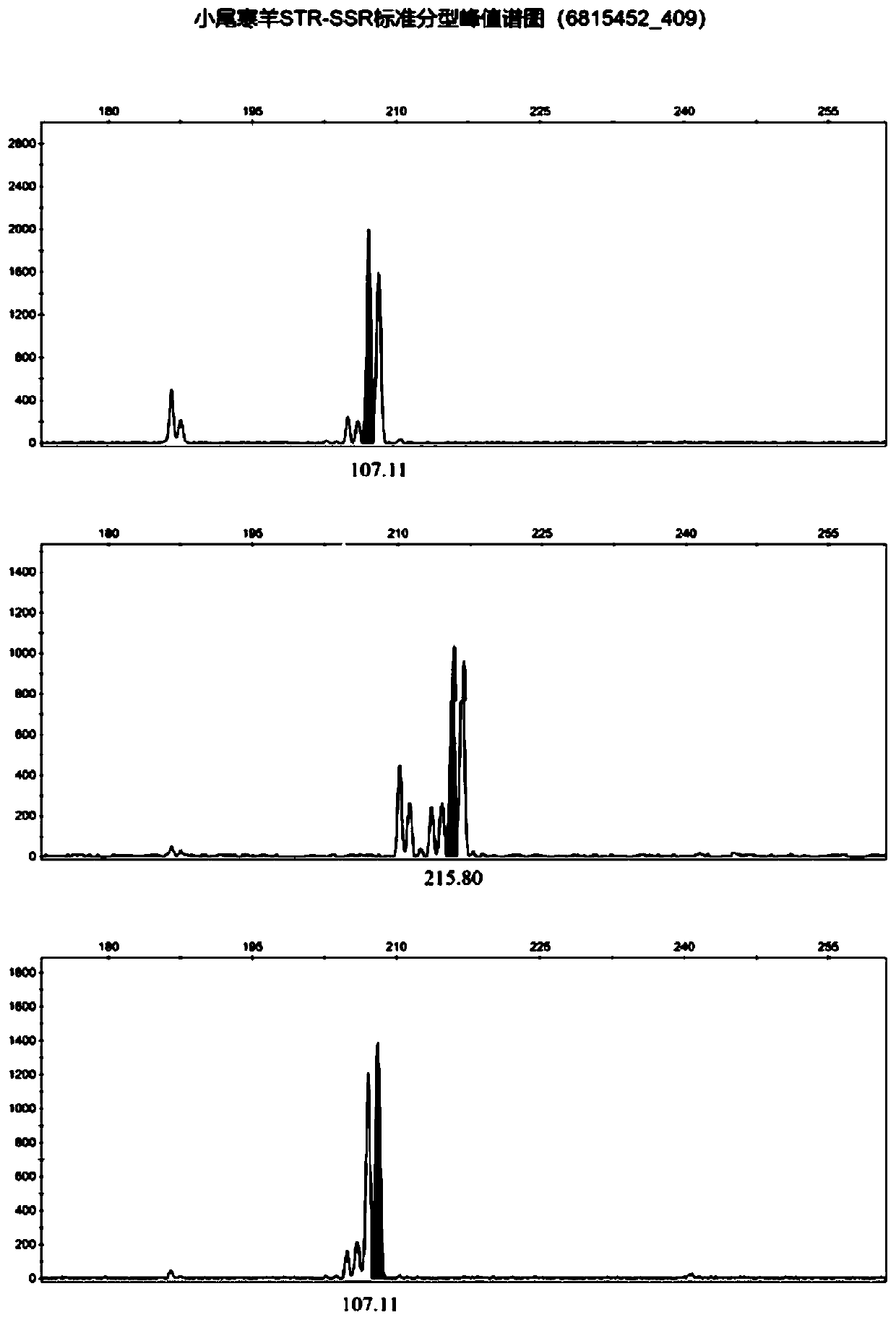
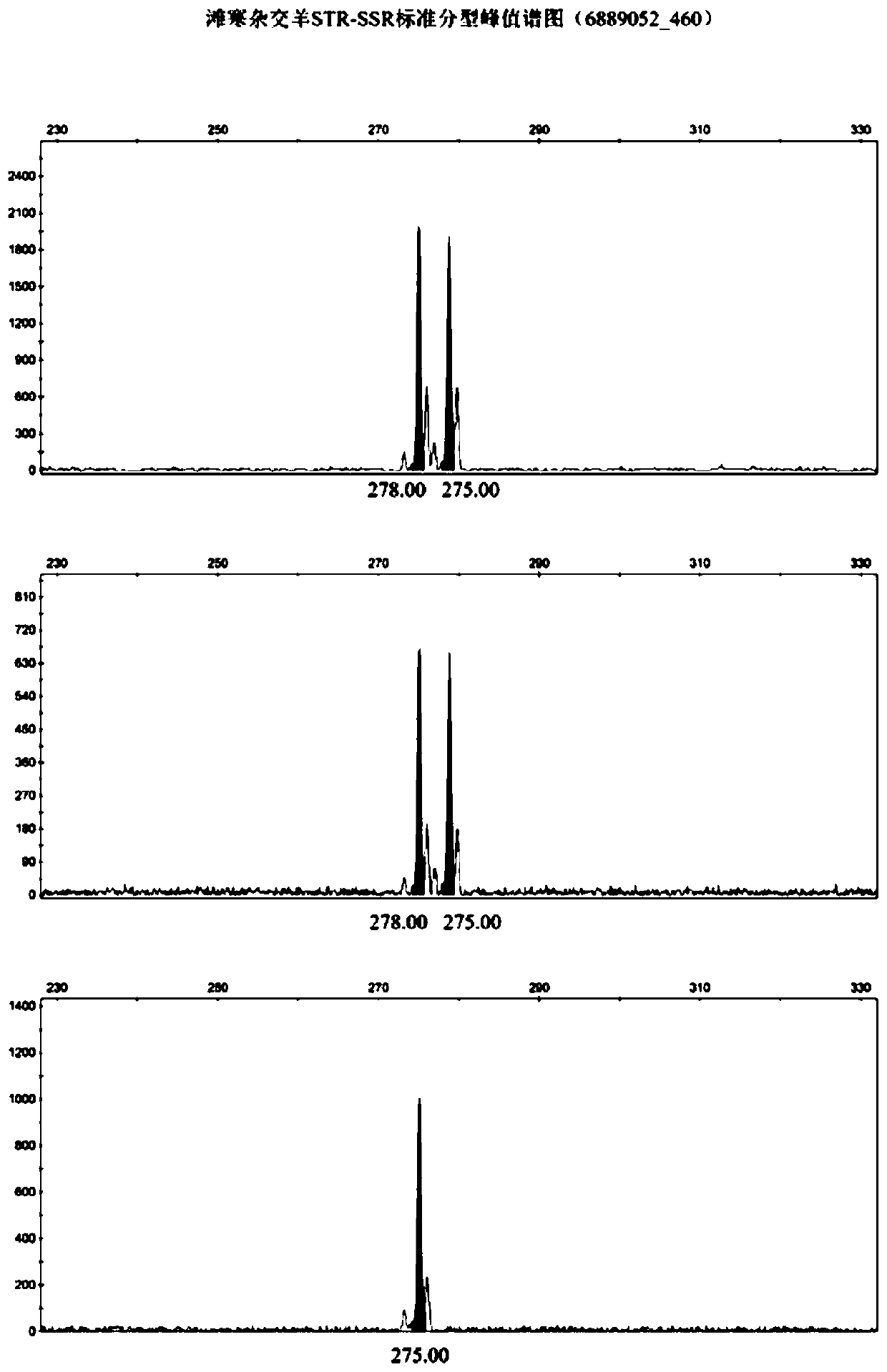
Affinis sheep meat detection method based on species specificity STR-SS
A detection method and specific technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, genetic engineering, etc., can solve the problems of long detection time, difficult identification, high cost, etc., and achieve simple and easy operation , time-consuming effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The first step: DNA extraction of standard mutton and mutton samples to be tested: take the muscle tissues of 25 Tan sheep, small-tailed Han sheep and Tanhan hybrid sheep, grind them into powder with liquid nitrogen, add them to a 1.5 μL centrifuge tube, and add 400 μL BufferDigestion , shake to mix. Water bath at 65°C for 1 hour until the cells are completely lysed; add 200 μL Buffer PA, mix thoroughly by inversion, and place in a refrigerator at -20°C for 5 minutes; centrifuge at 10,000 rpm at room temperature for 5 minutes, transfer 500 μL of the supernatant to a new 1.5 μL centrifuge tube Put it into a centrifuge and centrifuge at 12,000rpm to take the supernatant; add an equal volume of isopropanol, invert it 5 times to make it fully mixed, and let it stand at room temperature for 2min; centrifuge at 10,000rpm at room temperature for 5min, discard the supernatant; Rinse upside down for 1 min, centrifuge at 10,000 rpm for 2 min, discard the supernatant; open the lid...
Embodiment 2
[0069] The first step: DNA extraction of standard mutton and mutton samples to be tested: take the muscle tissues of 40 Tan sheep, small-tailed Han sheep and Tanhan hybrid sheep, grind them into powder with liquid nitrogen, add them to a 1.5 μL centrifuge tube, and add 400 μL BufferDigestion , shake to mix. Water bath at 65°C for 1 hour until the cells were completely lysed; add 200 μL Buffer PA, mix thoroughly by inversion, and place in a refrigerator at -20°C for 5 minutes; centrifuge at 10,000 rpm at room temperature for 5 minutes, transfer 525 μL of the supernatant to a new 1.5 μL centrifuge tube Put it into a centrifuge and centrifuge at 12,000rpm to take the supernatant; add an equal volume of isopropanol, invert it 7 times to make it fully mixed, and let it stand at room temperature for 2.5min; centrifuge at 10,000rpm at room temperature for 5min, discard the supernatant; Ethanol, rinse upside down for 2 minutes, centrifuge at 10,000rpm for 2 minutes, discard the supern...
Embodiment 3
[0077] The first step: DNA extraction of standard mutton and mutton samples to be tested: take the muscle tissues of 50 Tan sheep, small-tailed Han sheep and Tanhan hybrid sheep, grind them into powder with liquid nitrogen, add them to a 1.5 μL centrifuge tube, and add 400 μL BufferDigestion , shake to mix. Bath at 65°C for 1 hour until the cells are completely lysed; add 200 μL Buffer PA, mix thoroughly by inversion, place in a refrigerator at -20°C for 5 minutes; centrifuge at 10,000 rpm at room temperature for 5 minutes, transfer 550 μL of the supernatant to a new 1.5 μL centrifuge tube Put it into a centrifuge and centrifuge at 12,000rpm to take the supernatant; add an equal volume of isopropanol, invert it 8 times to make it fully mixed, and let it stand at room temperature for 3min; centrifuge at room temperature for 5min at 10,000rpm, discard the supernatant; Rinse upside down for 3 minutes, centrifuge at 10,000rpm for 2 minutes, discard the supernatant; open the lid an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com