A kind of method for breeding high-yield Wenguan fruit line
A high-yielding technology for sorbifolium sorbifolium, applied in the fields of botany equipment and methods, biochemical equipment and methods, and microbial measurement/inspection, can solve the problem of the lack of methods for identifying self-fertile, high-yielding and superior plants of sorbifolium sorbifolium New varieties breeding technology and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Embodiment 1, the acquisition of X. sorbifolium genomic DNA
[0064] The genomic DNA of the sample was extracted by the CTAB method, and the specific operation was as follows:
[0065] 1) Preheat the CTAB extract in a 65°C water bath;
[0066] 2) Grind the sample quickly in liquid nitrogen, transfer the powdered material into a 2mL centrifuge tube, add preheated CTAB extract (3-5ml of extract per gram of sample), and keep warm at 65°C for 30-60min, every Gently invert and mix for 10 minutes;
[0067] 3) Centrifuge at 11000rpm for 5min, take the supernatant and transfer it to a new centrifuge tube;
[0068] 4) Add an equal volume of phenol / chloroform (1:1, volume ratio), mix thoroughly, centrifuge at 11,000 rpm for 10 min, and transfer the supernatant to a new centrifuge tube;
[0069] 5) Add an equal volume of chloroform, mix thoroughly, centrifuge at 11000rpm for 10min, and transfer the supernatant to a new centrifuge tube;
[0070] 6) Repeat steps 4) and 5);
[0...
Embodiment 2
[0076] Embodiment 2, SSR primer screening
[0077] (1) PCR reaction system:
[0078] SSR (total 20 μL): ddH 2 7.2 μL of O, 10 μL of MIX, 0.3 μL of forward primer F, 0.3 μL of reaction primer R, 2 μL of DNA template (genomic DNA of X. sorbifolium obtained in Example 1), and 0.2 μL of Taq.
[0079] (2) The PCR reaction adopts the following cycle parameters:
[0080] SSR PCR amplification program: pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 s, annealing at 54°C for 35 s, extension at 72°C for 40 s, a total of 35 cycles; final extension at 72°C for 3 min.
[0081] (3) Primer information:
[0082] Table 1 Screening primer information
[0083]
[0084]
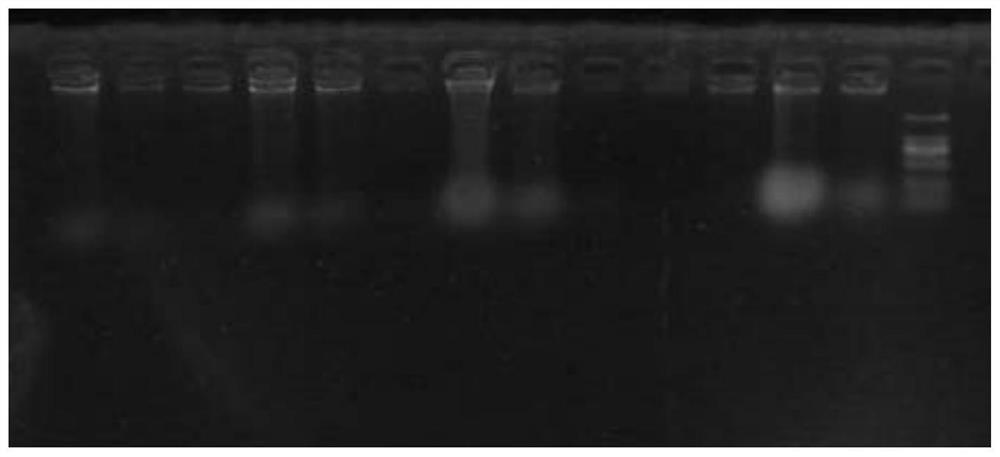
[0085] (4) The PCR product was added with loading buffer, denatured at 94° C. for 10 min, then analyzed by vertical electrophoresis on a 6% denaturing polyacrylamide gel, and observed after silver staining. Partial picture of primer screening in denaturing polyacrylamide gel electrophoresis (PAGE) ...
Embodiment 3
[0086] Embodiment 3, detection by capillary electrophoresis
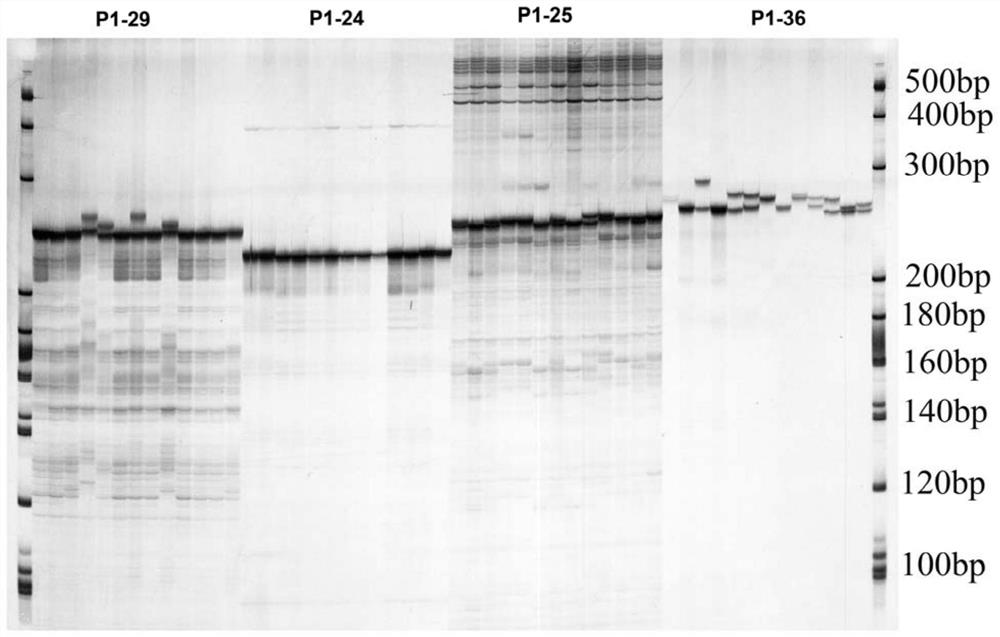
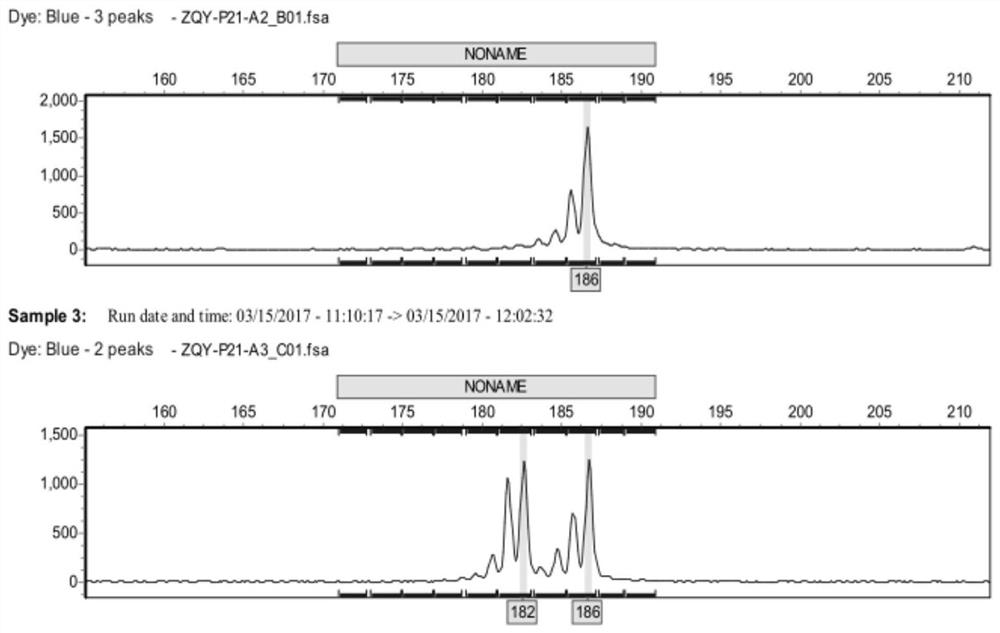
[0087] Using the 19 pairs of SSR primers (Table 1) with clear bands, high polymorphism and good repeatability screened in Example 2 above, fluorescent primers were synthesized, and the fragment size was detected by capillary electrophoresis analysis.
[0088] 1. PCR amplification
[0089] (1) PCR reaction system:
[0090] SSR fluorescent primer system (total 20 μL): ddH 2 O 14.8 μL, dNTP 0.4 μL, Buffer 2 μL, forward primer F 0.3 μL (20 μM), reverse primer R 0.3 μL (20 μM), DNA template 2 μL, Taq 0.2 μL.
[0091] (2) The PCR reaction adopts the following cycle parameters:
[0092] SSR PCR amplification program: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 54°C (annealing temperature fluctuates around 54°C) for 35 s, extension at 72°C for 40 s, a total of 35 cycles; final extension at 72°C for 3 min.
[0093] 2. Capillary electrophoresis analysis
[0094] After mixing the formam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com