S-adenosylmethionine synthetase mutant and preparation method thereof
A technology of adenosylmethionine and synthase, which is applied in the field of S-adenosylmethionine synthase mutant and its preparation, and can solve the problems of low enzyme catalysis efficiency, ineffective accumulation, low bioavailability and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Construction of S-adenosylmethionine synthetase recombinant bacteria
[0025] Select the S-adenosylmethionine synthetase derived from Escherichia coli K12, use PCR technology, use the E.coli K12 genome as a template, and use metK-F:5'-GATCC GAATTC ATGGCAAAACACCTTTTTACG-3' (SEQ ID NO.5) and metK-R:5'-GTG CTCGAG TTACTTCAGACCGGCAGCAT (SEQ ID NO.6) was used as a primer to amplify the metK gene, and EcoR I and Xho I restriction endonuclease sites (underlined) were introduced at its 5' end and 3' end respectively. The PCR reaction system (50 μL) was: 25 μL of 2×PrimeSTAR Max Premix, 1 μL of template DNA, 1 μL of upstream and downstream primers, and 22 μL of sterile water. The PCR reaction conditions were: pre-denaturation at 98°C for 5 min; denaturation at 98°C for 10 s, annealing at 60°C for 5 s, extension at 72°C for 10 s, and 30 cycles; extension at 72°C for 5 min. Verify the PCR amplification product with 1% agarose gel electrophoresis, when the target band...
Embodiment 2
[0026] Example 2: Single point mutation of S-adenosylmethionine synthetase gene
[0027] After culturing the recombinant strain E.coli BL21(DE3) / pET-28a(+)-metK in Example 1 overnight, extract the recombinant plasmid pET-28a(+)-metK, and use it as a template to fix the site at position 303 mutation. The FastMutagenesis system kit was used for site-directed mutagenesis, and I303V-F: 5'-TCAGGTTTCCTACGCAGTCGGCGTGGC-3' (SEQ ID NO.7) and I303V-R: 5'-CCAGAAGGCACTTTCTCAGTACCGAAAG-3' (SEQ ID NO.8) were used as primers to carry out the whole Plasmid PCR amplification (94°C 5min; 94°C 20s, 60°C 20s, 72°C 60s, 30 cycles; 72°C 10min). After the PCR product was verified by agarose gel electrophoresis, the template was digested with the restriction enzyme DpnI, and then the digested product was transferred into E.coli BL21(DE3) competent cells by heat shock method to obtain a single point mutation recombinant bacterium . The amino acid sequence of the single point mutation is SEQ ID NO.1...
Embodiment 3
[0028] Example 3: Construction of two point mutations in S-adenosylmethionine synthetase
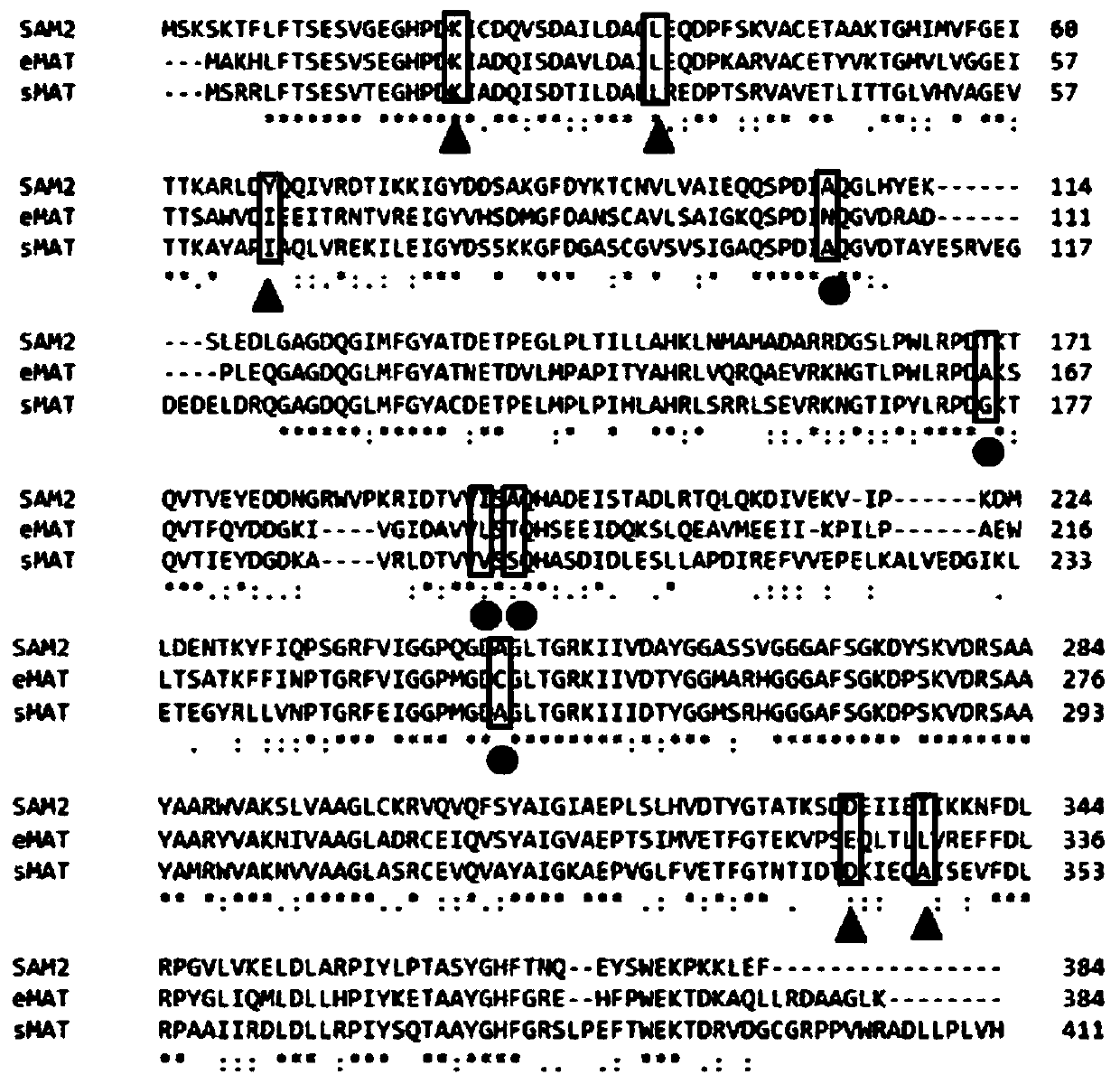
[0029] The amino acid sequences of eMAT, SAM2 and sMAT were homologously compared, and the corresponding mutation sites in eMAT and pDS16 and pDS56 were found, and the points that could be mutated were: K18R, L31P, I65V, E324G, L329V( figure 2, marked with a triangle). Using the mutant I303V coding gene as a template, K18R-F:5'-TCTGAAGGGCATCCTGACAGAATTGCTGAC-3'(SEQ ID NO.9) and K18R-R:5'-CTGTCAGGATGCCCTTCAGAGACGGAC-3'(SEQ ID NO.10); I31P-F :5'-TGATGCCGTTTTAGACGCGCCCCTCGAACAG-3'(SEQ ID NO.11) and I31P-R:5'-GGCGCGTCTAAAACGGCATCAGAAATTTGG-3'(SEQ ID NO.12); I65V-F:5'-ACCAGCGCCTGGGTAGACGTCGAAGAGATC-3'(SEQ ID NO. .13) and I65V-R: 5'-CGTCTACCCAGGCGCTGGTGGTGATTTCG-3' (SEQ ID NO. 14); E324G-F: 5'-TACTGAGAAAGTGCCTTCTGGACAACTGACC-3' (SEQ ID NO. 15) and E324G-R: 5'-CCAGAAGGCACTTTTCAGTACCGAAAG -3' (SEQ ID NO.16); L329V-F:5'-TTCTGAACAACTGACCCTG GTG GTACGTGAG-3' (SEQ ID NO.17) and L329V-R:5'-CCAGG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com