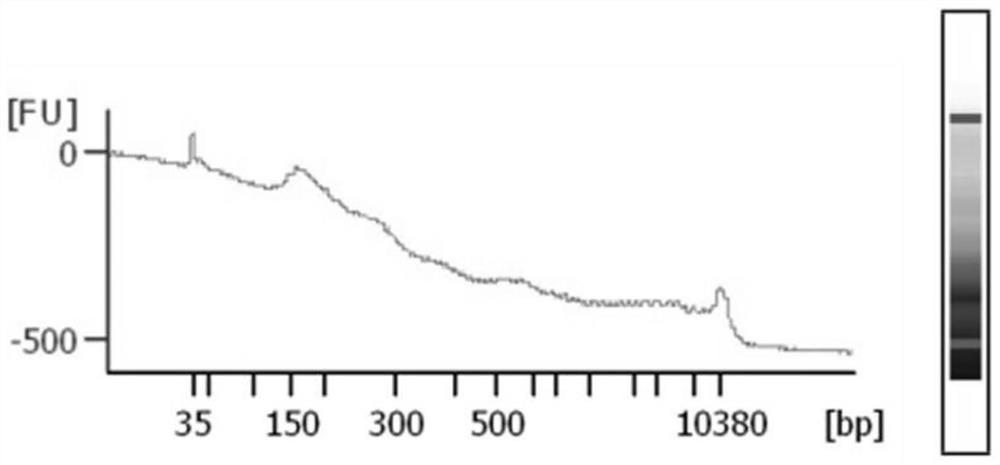
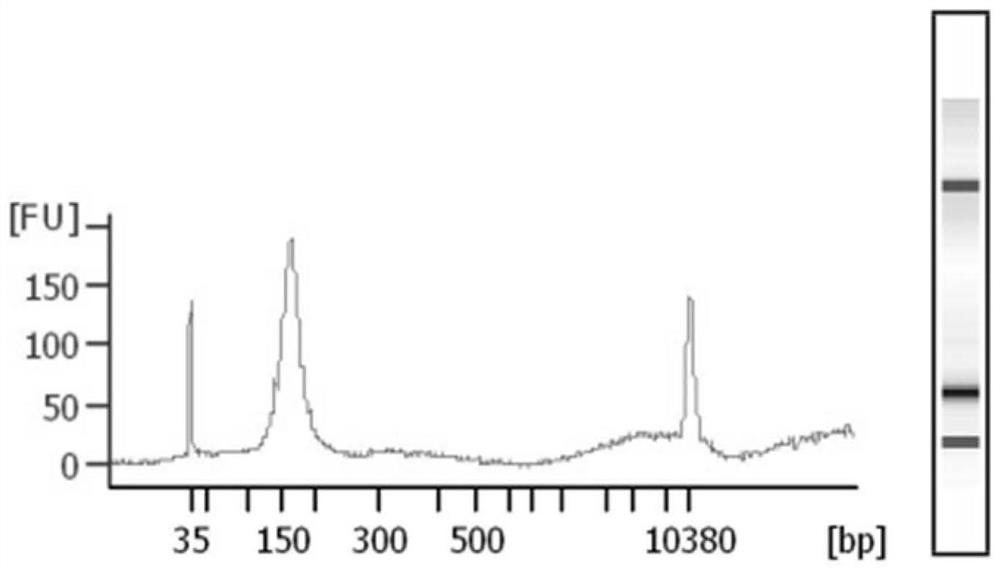
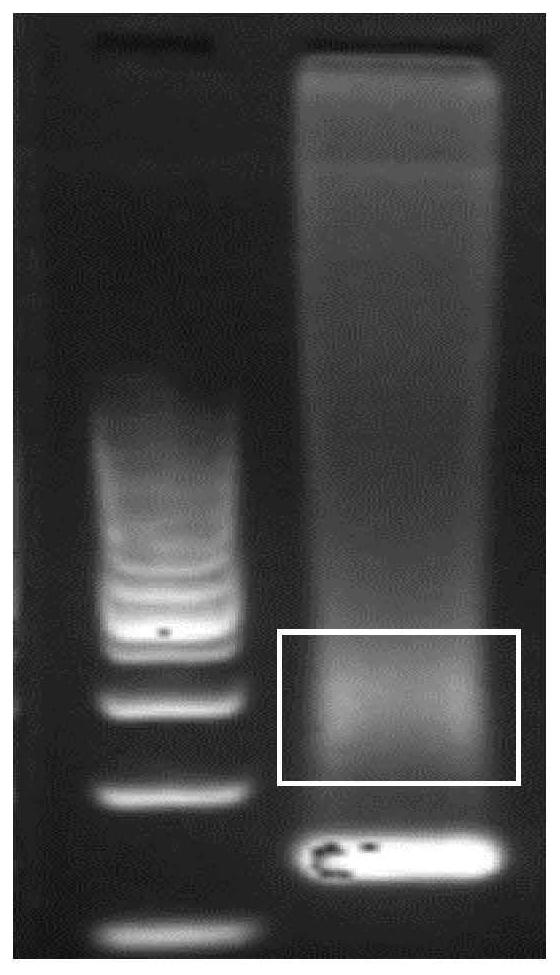
A method for extracting cfDNA from follicular fluid
A technology of follicular fluid and lysate, which is applied in the field of genetic testing, can solve the problems of many fragments, less research, and complex components of follicular fluid, and achieve the effects of saving time and cost, simplifying extraction steps, and reducing waste
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Extraction of cfDNA
[0039] 1. Separation and preservation of follicular fluid
[0040]The centrifuge was pre-cooled at 4°C, centrifuged at 3000g for 15min, and the supernatant was carefully aspirated. The supernatant was then centrifuged at 16,000g for 10 min, and the supernatant was transferred to a new centrifuge tube. It can be stored at 4°C if used on the same day, or at -80°C if stored for a long time.
[0041] 2.1, configure the lysate
[0042] 100 mM Tris-HCl, 20-30 mM EDTA, 500 mM NaCl, 1% SDS, along with 6 μg Carrier RNA per ml, vortex to mix.
[0043] 2.2, cracking
[0044] Take a new 15ml centrifuge tube, add 100μl proteinase K (1mg / ml), then add 1ml follicular fluid and 800μl lysate in sequence, vortex at maximum speed for 30s, and incubate the centrifuge tube at 60°C for 30min.
[0045] 3. Adsorption of cfDNA by carboxyl magnetic beads
[0046] Add 0.5ml of isopropanol to the above centrifuge tube, after vortexing for 30s, add 200μl carbox...
Embodiment 2
[0060] Example 2 Construction of high-throughput sequencing library
[0061] (1) cfDNA end repair
[0062] Add the corresponding samples and reagents to the PCR tube according to the reaction system in Table 1, vortex to mix well, and then put them in the PCR machine, and incubate at 20 °C for 5 min. After the reaction is over, add ddH 2 O to the final volume of the system 200 μl. Then transfer it to a new 1.5ml centrifuge tube, add 200μl of mixed phenol (phenol:chloroform:isoamyl alcohol=25:24:1, pH=8.0) to the system (the same volume as the sample to be purified), shake vigorously Mix well and centrifuge at 13000rpm for 5min at room temperature. Transfer the supernatant to a new 1.5ml tube (be careful not to aspirate the protein layer at the junction of the aqueous phase and the organic phase at this time to avoid contamination of genomic DNA by aspirated protein. It is not necessary to completely absorb the aqueous phase, and leave a little to avoid contamination). Add ...
Embodiment 3
[0085] Example 3 High-throughput sequencing
[0086] High-throughput sequencing of the high-throughput sequencing library of cfDNA in Example 2 was performed using Illumina Hiseq 2000. The sequencing data was used for basic quality inspection using Fastqc software, and the data quality was qualified before further use. The PCR primer sequences used for sequencing are shown in Table 6.
[0087] Table 6 PCR primer sequences
[0088]
[0089] The quality inspection results of paired-end sequencing are as follows: Figure 5 As shown, most of the base quality is above 30, indicating that the data quality is good. The meaning of the bar is 25%-75% of the data distribution, and only the data whose total quality score is above 30 (representing a sequencing error rate of 1 in 1,000) can be further analyzed.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com