SSR molecular marker for screening content and phenotype of apple fruit sugar and application thereof
A technology of molecular markers and sugar content, applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of high heterozygosity, restriction of breeding process, and long childhood in apples, and achieve efficient screening Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Detection of sugar accumulation-related genes by SSR molecular marker typing
[0027] (1) Determination of population phenotype data
[0028] Ripe apple fruits were picked and their skins and cores were removed, and the apple pulp cut into small pieces was quick-frozen in liquid nitrogen and stored in a -80°C ultra-low temperature refrigerator. Grind the pulp sample frozen in a -80°C ultra-low temperature refrigerator into a powder, weigh 1g of the pulp powder sample into a 15mL centrifuge tube and add 6mL ultrapure water to fully mix the sample. Centrifuge in the machine for 15min at a speed of 5000rpm. The supernatant produced by centrifugation was filtered through a 0.22 μm Sep-Pak microporous membrane, and the filtrate was saved for sugar content determination. The chromatographic conditions for sugar determination are: CarboSep CHO-620 calcium sugar column (300×6.5mm, pore size 10μm) and CarboSep CHO-620 guard column core, the operating temperature is ...
Embodiment 2
[0044] Example 2: Verification of Candidate Gene Functions in Strawberry Fruit and Apple Callus Systems
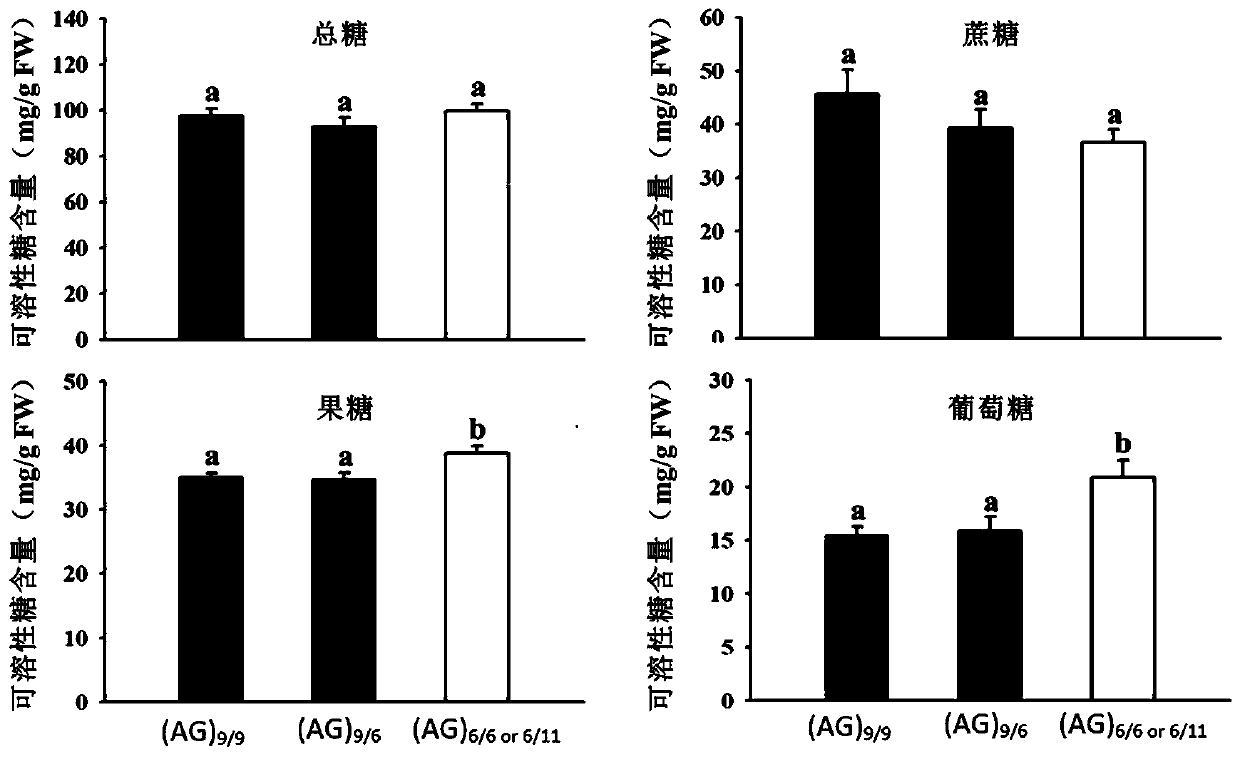
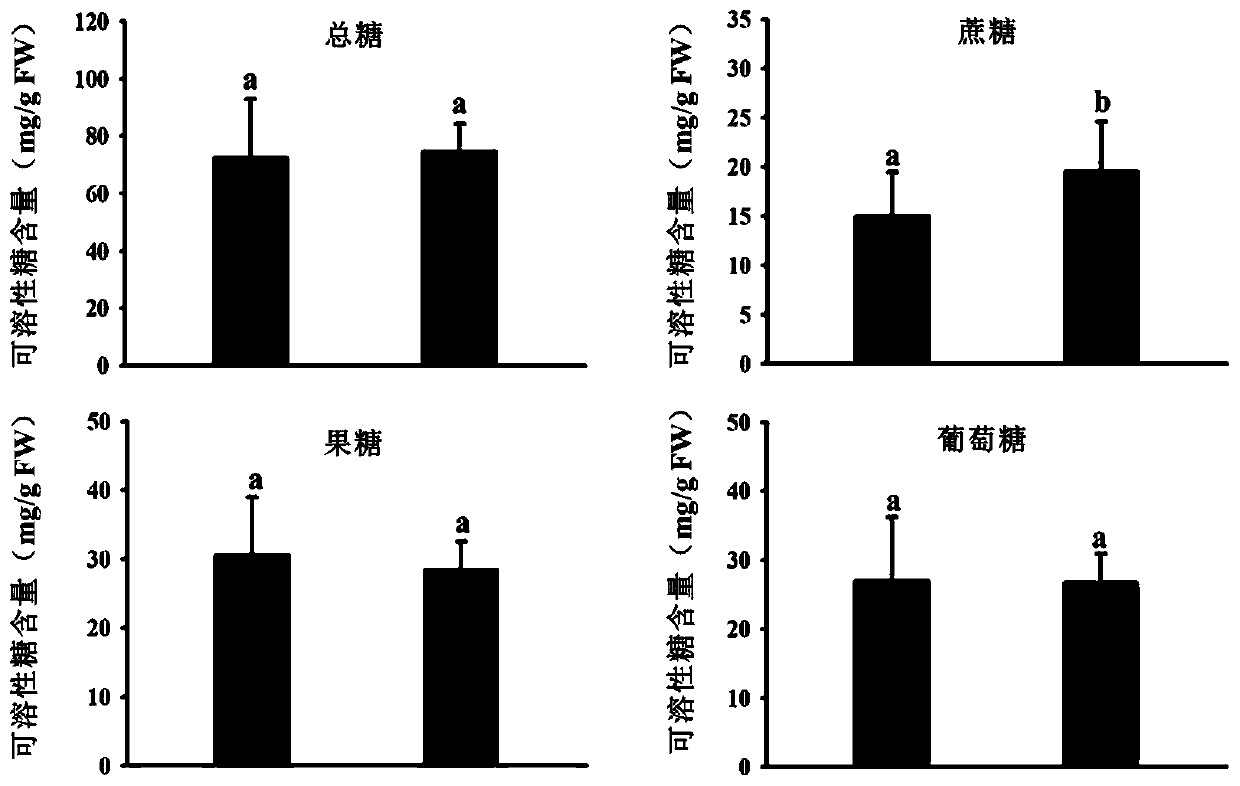
[0045] The coding sequence of the MdSUT4.1 gene corresponding to the SSR molecular marker SugarA3 was inserted into the multiple cloning site of the pSAK277 binary vector to construct the pSAK277-MdSUT4.1 expression vector and transformed into EHA105 Agrobacterium competent cells by heat shock method. The strains were cultured in LB liquid medium containing corresponding antibiotics in a shaker at 28° C. to OD600 = 1.0, and the bacterial cells were collected to prepare an Agrobacterium infection solution. Agrobacterium infection solution A (5mg / mL glucose, 50mM MES, 2mM Na 3 PO 4 12H 2 0 and 100 μM acetosyringone) to resuspend the bacterial body to OD600=0.2, inject the bacterial solution from the base of the green fruit of "Hongyan" strawberry through a 1mL syringe, place the transformed strawberry plants in a 16°C greenhouse for 9 days, and pick the strawberries The t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com