Indel molecular marker for identifying novel single fasciated inflorescence of capsicum annuum L., primer and application
A molecular marker and inflorescence technology, applied in the field of pepper breeding and molecular biology, can solve the problems of low selection efficiency, slow breeding progress and backwardness, and achieve the effects of shortening the breeding cycle, saving manpower and material resources, and reducing workload.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] In this embodiment, an Indel molecular marker for identifying a novel single-clustered inflorescence of pepper is designed based on the upstream and downstream sequences of insertions / deletions closely linked with the new-type pepper-clustered inflorescence gene.
[0048] The steps for obtaining the above-mentioned molecular markers closely linked with the pepper novel cluster inflorescence gene are as follows:
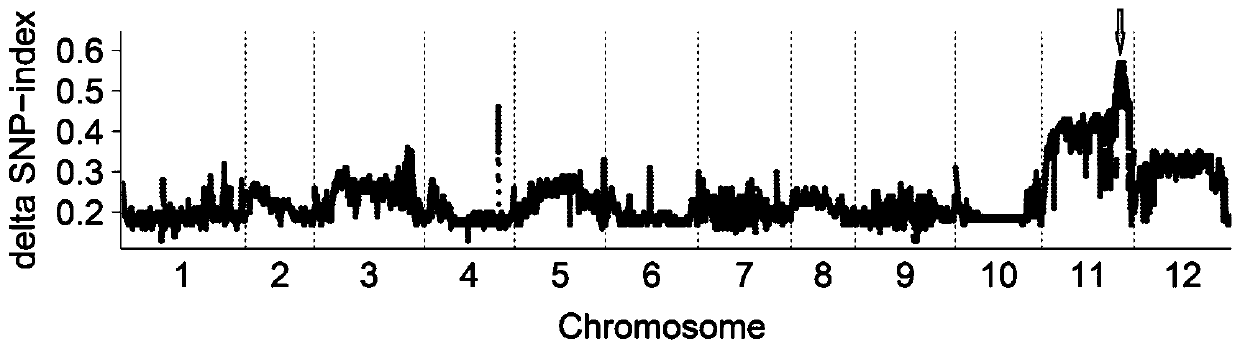
[0049] (1) Population construction: using pepper solitary inflorescence high-generation inbred lines L816 and CJ220 as female parents, and clustered inflorescence mutant CL74 as male parent, construct F2 populations CS and SY ( figure 1 ). CL74, L816 and CJ220 are all stored in the Pepper Germplasm Resource Bank of Hunan Vegetable Research Institute and can be sold;
[0050] (2) Phenotype identification: two F2 populations, CS and SY, were planted in a glass greenhouse for conventional cultivation and management. The phenotype identification of the individua...
Embodiment 2
[0059] Validation of tightly linked molecular markers for novel cluster inflorescence genes in pepper:
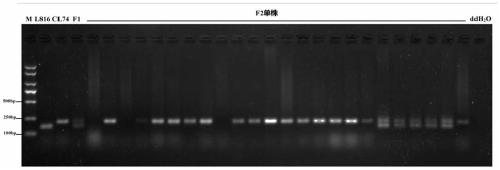
[0060] Using the molecular markers obtained in Example 1 to identify 50 clustered recessive individuals randomly selected from the two F2 populations of CS and SY, the steps are as follows:
[0061] (1) Use the CTAB method to extract the DNA of a single plant of pepper F2 population as a template, the DNA concentration is 30-100ng / ul, and use molecular markers for PCR amplification. The forward primer sequence is CaDH-F: CGGGAGGCTATGTGACATTC, and the reverse primer sequence is CaDH -R: TCTACGTCGTCCACGTTCAA, PCR reaction system: the total volume is 20μl, and the specific components are as follows: PCR is carried out in a 20μL reaction system consisting of 1μl genomic DNA (30ng / μl), 18μl PCR-T3-Mix (TSINGKE) and forward and reverse primers (both primer concentrations are 10 μmol) each consisting of 0.5 μl;
[0062] (2) The PCR reaction was carried out on the S1000 PCR machin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com