Method for differentiating 5-methylcytosine and 5-hydroxymethylcytosine in DNA
A technology for hydroxymethylated cytosine and methylated cytosine, which is applied in the field of distinguishing 5-methylated cytosine and 5-hydroxymethylated cytosine in DNA, and can solve the problem of large sequence preference and low transformation efficiency and other problems, to achieve the effect of improving timeliness, low damage rate, and increasing stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1, expression and purification of APOBEC3A enzyme
[0045] This application uses pET41a as the vector framework, amplifies the MBP-Xa tag from pMal-c5x, transfers it to pET41a through homologous recombination, and then connects the APOBEC3A gene and the vector to obtain the recombinant plasmid pET41a-MBP-A3A-His. Transform E.coli BL21(DE3)pLysS competent cells (Tiangen Biochemical) with the recombinant plasmid, and pick 10-20 small positive clones into 5mL liquid LB medium (containing 2.5mM glucose, 50ug / mL Kanamycin in chloramphenicol, 34ug / mL chloramphenicol, 100uM EDTA), 37°C, 250rpm to OD600 of about 0.5, quickly cooled on ice, then added IPTG to a final concentration of 0.2mM, and cultured overnight at 16°C. Collect the bacteria by centrifugation, resuspend the bacteria in 0.5ml PBS, add lysozyme and incubate at 37°C for 30min, and take the supernatant for Western Blot analysis. The clones whose expression was confirmed by WB were scaled up and cultured...
Embodiment 2
[0046] Example 2, verification of the deamination activity of APOBEC3A
[0047] 1. DNA substrate preparation: Amplify a DNA fragment from plasmid DNA by nested PCR, 580bp in length;
[0048] 2. APOBEC3A deamination conversion process
[0049] (1) Prepare APOBEC3A Enzyme Mix on ice according to the following system, shake slightly to mix evenly, centrifuge briefly and insert on ice for later use.
[0050] Table 1
[0051]
[0052]
[0053] (2) DNA denaturation: Mix the components according to the following system, insert on ice to cool for 1 min, transfer to a 95°C PCR instrument for denaturation for 3 min, quickly place in an ice-water bath, and let stand for 2 min.
[0054] Table 2
[0055]
[0056] (3) Quickly mix the Enzyme Mix and the denatured product, gently blow and mix with a pipette gun, centrifuge briefly at 4°C and place on ice for later use.
[0057] (4) Transfer the above-mentioned mixed product to a PCR instrument to start the transformation program....
Embodiment 3
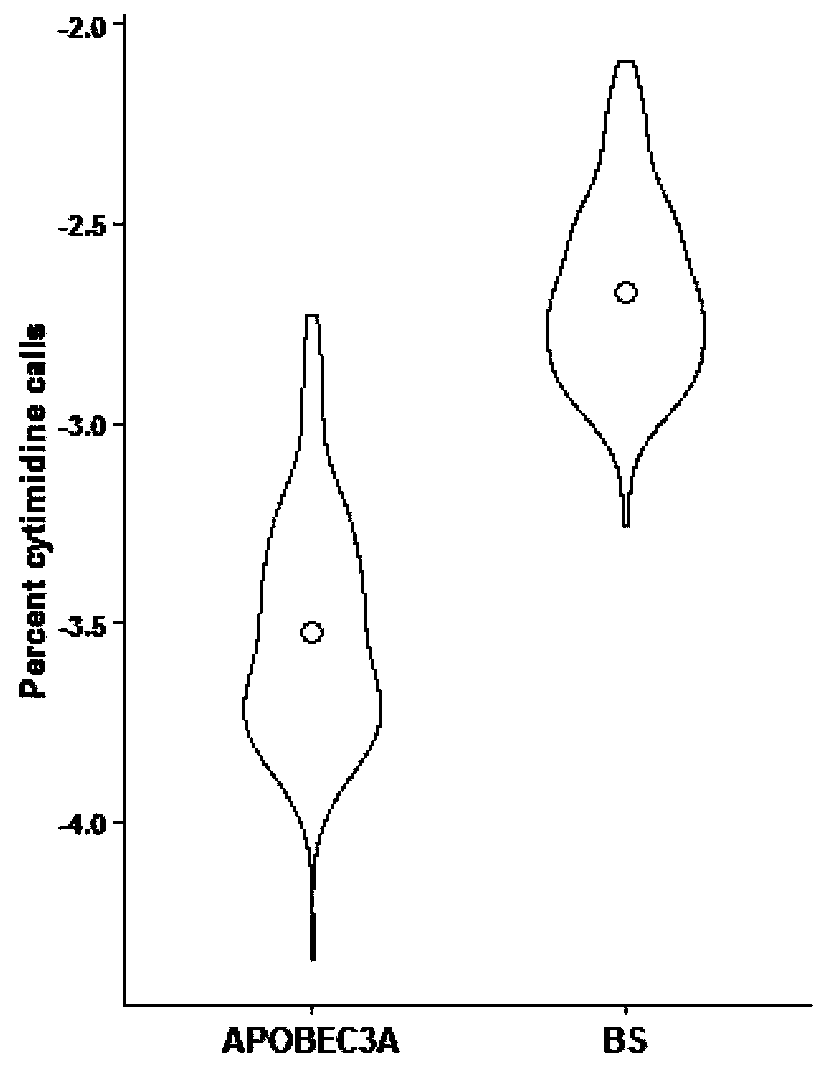
[0067] Example 3, using APOBEC3A to distinguish 5-mC and 5-hmC
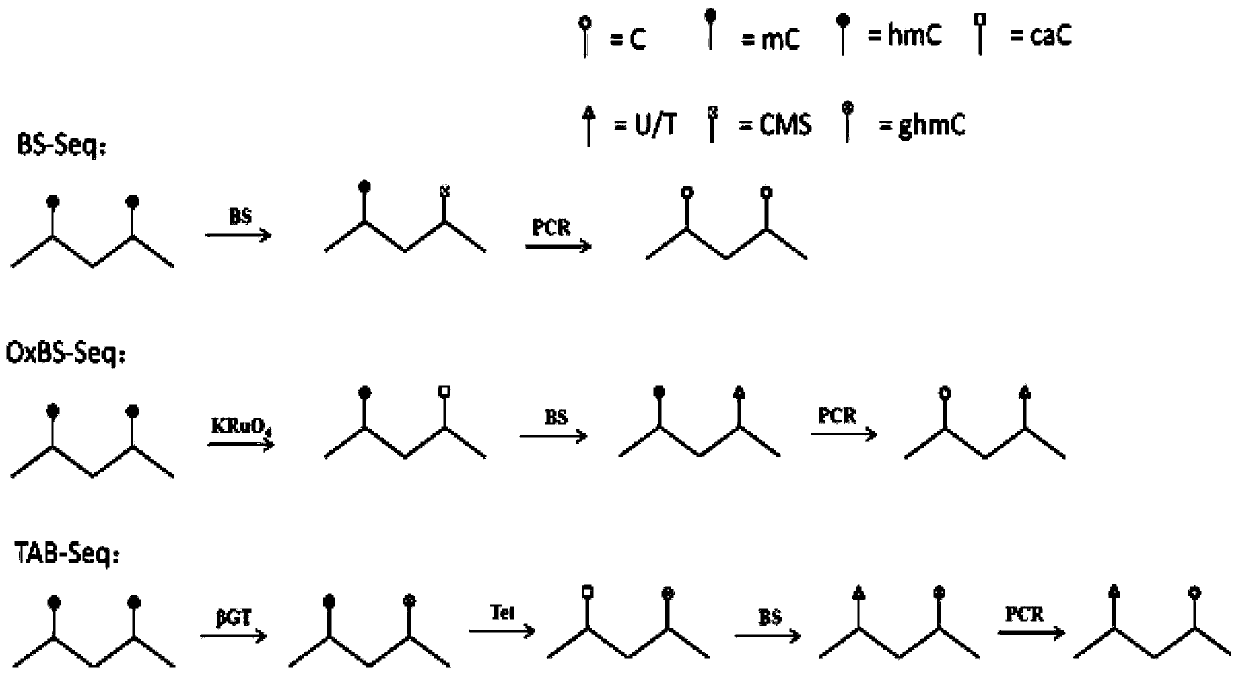
[0068] This example provides a method for quickly distinguishing 5-mC and 5-hmC based on the difference of APOBEC3A in the deamination conversion of 5-mC and 5-hmC. The principle is as follows Figure 4 shown.
[0069] 1. DNA substrate preparation
[0070] Use 5-mC dCTP instead of dCTP for PCR amplification, a DNA fragment amplified from λDNA is named mC-λ3, and a DNA fragment amplified from pMal-c5x_RH2 is named mC-RH2; use 5-hmC dCTP instead of dCTP PCR amplification, a DNA fragment amplified from λDNA was named hmC-λ3, and a DNA fragment amplified from pMal-c5x_RH2 was named hmC-RH2.
[0071] 2. Glycosylation of 5-hmC
[0072] Table 4 reaction system
[0073]
[0074]
[0075] Reaction conditions: add each component in order, mix well, transfer to PCR machine, incubate at 37°C for 1h, hold at 4°C, heat cover at 50°C.
[0076] Product recovery: After the reaction, use Oligo Clean&Concentrator (Zymo R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com