3-hydroxy-3-methylglutaryl coenzyme A reductase gene RKHMGR and application thereof
A technology of methylglutaryl coenzyme and reductase, which is applied to the invention belongs to the field of biotechnology and genetic engineering, and can solve the problems that the yield and quality of carotenoids cannot meet market demand and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1: from Rhodosporidium yeast ( Rhodosporidium kratochvilovae The nucleotide sequence of the 3-hydroxy-3-methylglutaryl-CoA reductase gene RKHMGR isolated from YM25235
[0019] The total RNA of Rhodosporidium YM25235 was extracted using the UNlQ-10 Column Trizol Total RNA Extraction Kit (product number: SK1321) of Sangon Bioengineering (Shanghai) Co., Ltd., and then the PrimeScript ® RT reagent of the TaKaRa company kit was used to extract the total RNA. Kit With gDNA Eraser (Perfect Real Time) was used to synthesize cDNA by reverse transcription, and 0.5 μL was used as a template for polymerase chain reaction. According to the sequence of RKHMGR found in transcriptome sequencing, specific primers RKHMGR-F and RKHMGR-R were designed ( Primer 1 and Primer 2), the cDNA template obtained above was used for PCR amplification on a PCR instrument (BIOER company), and the primers, reaction system and amplification conditions used in the reaction were as follows:
...
Embodiment 2
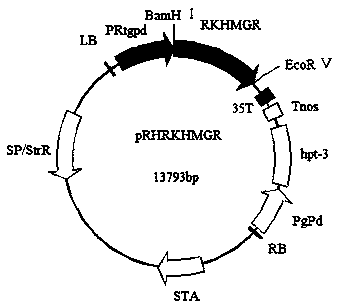
[0025] Example 2: Construction of overexpression vector pRHRHMGR
[0026] Using the reverse transcribed YM25235 cDNA as a template, using RKHMGR-F and RKHMGR-R as primers to amplify the coding sequence of RKHMGR, the obtained RKHMGR fragment size is about 4000bp, and the amplified RKHMGR fragment obtained by Bam H I , Eco RV two restriction endonucleases were digested, connected to the expression vector pRH2034 to obtain the recombinant plasmid pRHRKHMGR ( figure 2 ); the obtained recombinant plasmid was transferred into Escherichia coli DH5α for amplification, and after colony PCR verification, the recombinant plasmid was extracted and used Bam H I , Eco RⅤ carried out double digestion verification on pRHRKHMGR; the results showed that the recombinant plasmid pRHRKHMGR produced two bands of about 4 kb and 10.7 kb after double digestion ( image 3 Lane 3), these two bands are consistent with the size of the RKHMGR fragment and the fragment of the pRH2034 vector after d...
Embodiment 3
[0027] Example 3: Analysis of the relationship between RKHMGR gene and carotenoid synthesis in Rhodosporidium yeast
[0028] 1. Agrobacterium-mediated transformation of Rhodosporidium YM25235
[0029] The recombinant plasmid pRHRKHMGR was transformed into Rhodosporidium YM25235 by the Agrobacterium-mediated method, and the transformants were selected with the YPD medium containing Hygromycin B (Hygromycin B) at a final concentration of 150 μg / mL, and then followed by Shanghai Sangong Bioengineering Co., Ltd. DNA Extraction Kit instructions to extract the genomic DNA of yeast transformants, and then carry out PCR verification, the results are as follows Figure 4 shown.
[0030] 2. Analysis of RKHMGR gene and carotenoid content of Rhodosporidium yeast
[0031] After culturing the overexpressed strain containing pRHRKHMGR for 144 h at 28°C, the carotenoids were extracted, and the original Rhodosporidium YM25235 strain was used as a control, and the total carotenoid content was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com