Application of Arabidopsis mutant protoplasts in the analysis of signal transduction pathway
A technology for analyzing signals and protoplasts, applied in the direction of using vectors to introduce foreign genetic material, biochemical equipment and methods, recombinant DNA technology, etc., can solve the problems of no genome sequence, time-consuming and labor-intensive research on plant signal transduction pathways, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
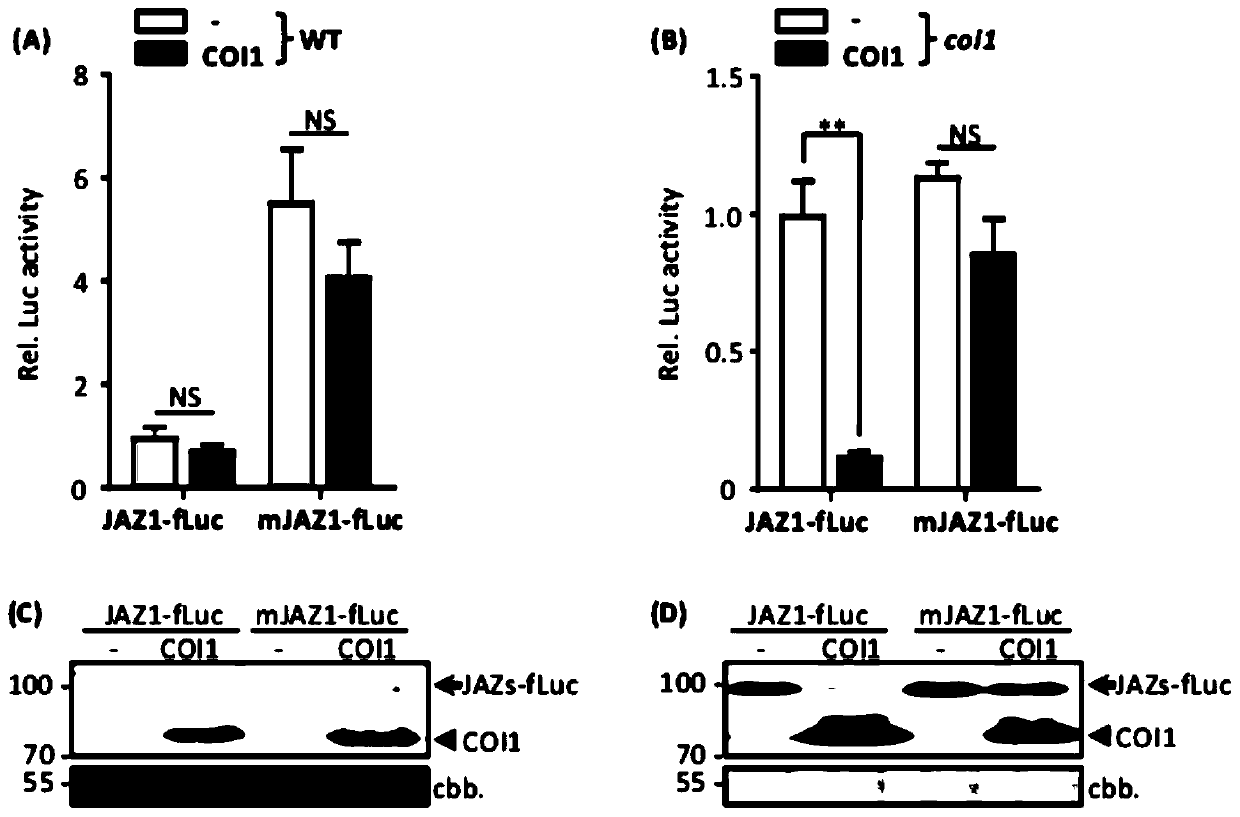
[0061] Example 1, JAZ1 protein degradation mediated by COI1 in Arabidopsis protoplast cells
[0062] 1. Construction of recombinant plasmids
[0063] 1. Construction of protein fusion reporter gene vector (i.e. recombinant plasmid UBQ10pro: HA-JAZ1-fLuc)
[0064] (1) The pBGWL7.0 vector was digested with restriction enzymes HindIII and XmaI, and a DNA fragment 1 of about 2303 bp was recovered. DNA fragment 1 contains the gene encoding firefly luciferase.
[0065] (2) The UBQ10pro:HA-GW vector was digested with restriction enzymes HindIII and XmaI, and the vector backbone 1 of about 10152 bp was recovered.
[0066] (3) Ligate the DNA fragment 1 and the vector backbone 1 to obtain the recombinant plasmid UBQ10pro: HA-GW-fLuc.
[0067] The nucleotide sequence of the recombinant plasmid UBQ10pro:HA-GW-fLuc is shown in SEQ ID No:1.
[0068] (4) Using the genomic DNA of wild-type Arabidopsis as a template, a primer pair consisting of JAZ1-gw-d1 and JAZ1ohnestop-gw-r1 was used fo...
Embodiment 2
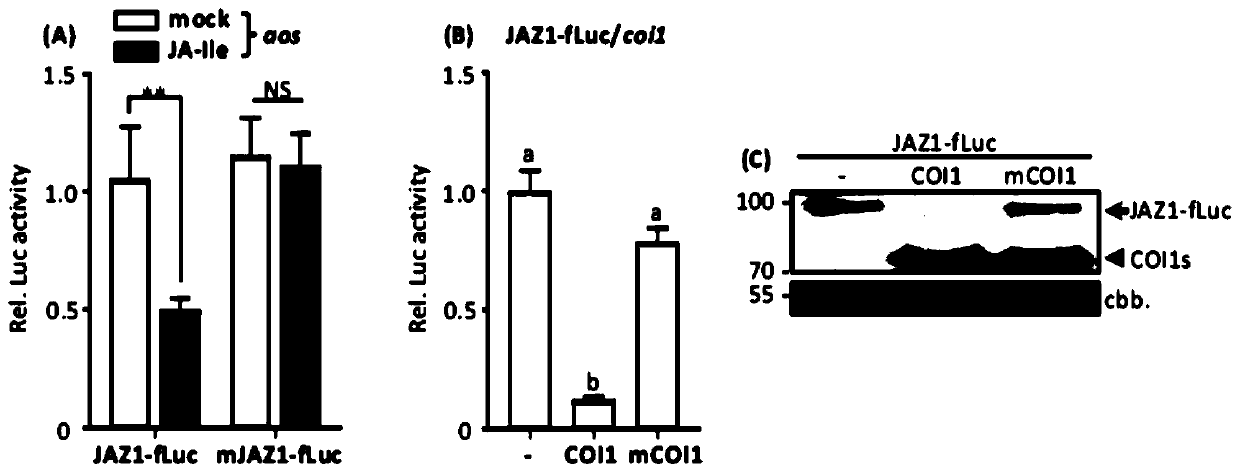
[0112] Example 2, JAZ1 protein degradation mediated by COI1 in protoplasts requires jasmonic acid
[0113] In Arabidopsis, endogenous JA molecules mediate the interaction between COI1 and JAZ. To determine whether JAZ1-fLuc proteolysis by COI1 in protoplasts is dependent on JA, the inventors of the present invention performed JA complementation experiments in protoplasts of aos mutants. The AOS gene, one of the key enzymes encoding the JA biosynthetic pathway, is mutated in the aos mutant, so JA cannot be synthesized.
[0114] 1. JA-Ile triggers the specific degradation of JAZ1 in protoplast cells
[0115] 1. Construction of recombinant plasmids
[0116] (1) The construction of the protein fusion reporter gene vector (ie, the recombinant plasmid UBQ10pro: HA-JAZ1-fLuc) is the same as step 1 in Example 1.
[0117] (2) The construction of the protein fusion reporter gene vector (ie, the recombinant plasmid UBQ10pro: HA-mJAZ1-fLuc) is the same as Step 1 and Step 2 of Example 1...
Embodiment 3
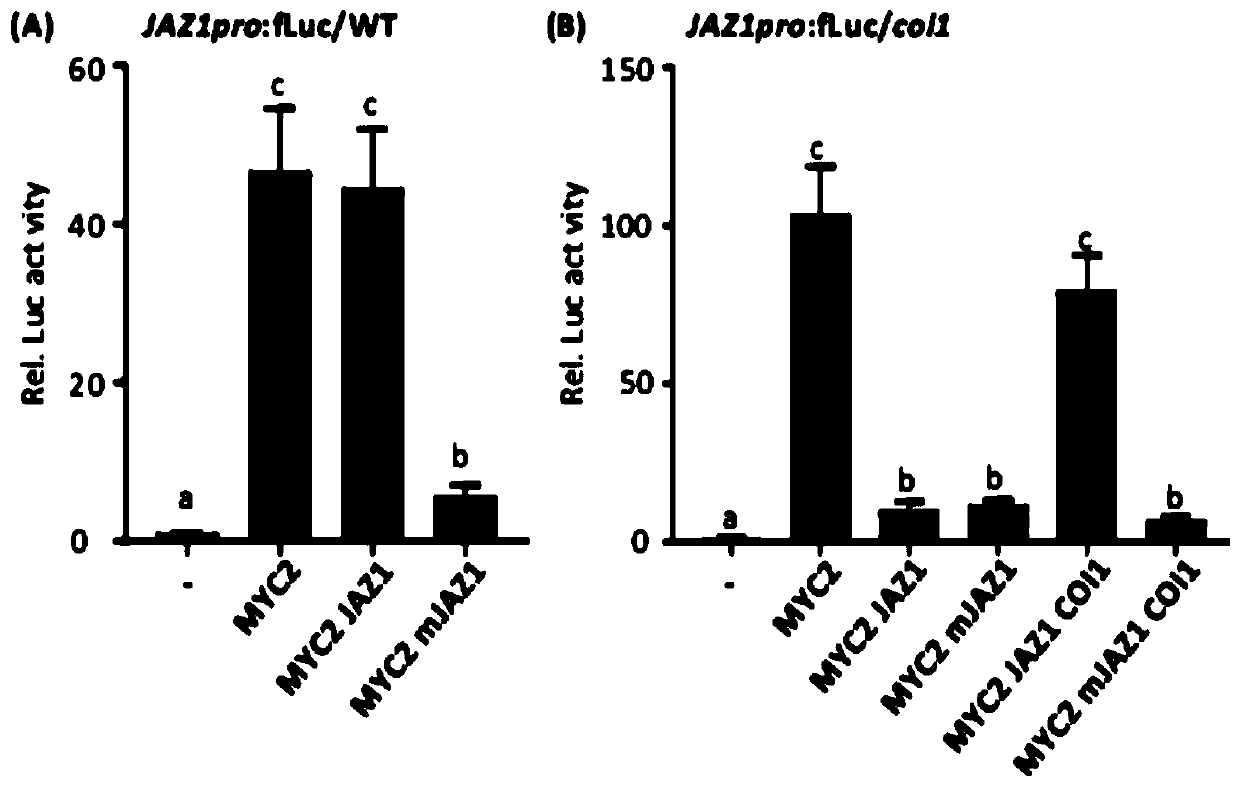
[0156] Example 3, COI1 releases JAZ1-inhibited MYC2 transcriptional activity
[0157] 1. Construction of recombinant plasmids
[0158] 1. Construction of recombinant plasmid JAZ1pro: fLuc
[0159] (1) Using the genomic DNA of wild-type Arabidopsis as a template, the primer pair composed of JAZ1pro-gw-d1 and JAZ1pro-gw-r1 was used for PCR amplification to obtain a promoter fragment of about 1267bp.
[0160] (2) The promoter fragment and the vector pDONR207 were subjected to BP reaction to obtain the intermediate vector pDONR207-JAZ1pro.
[0161] (3) The intermediate vector pDONR207-JAZ1pro and the pBGWL7.0 vector were subjected to LR reaction to obtain the recombinant plasmid JAZ1pro:fLuc.
[0162] In the recombinant plasmid JAZ1pro:fLuc, the gene encoding firefly luciferase is driven by the JAZ1 promoter.
[0163] 2. Construction of recombinant plasmid UBQ10pro: HA-JAZ1 (i.e. effector carrier JAZ1)
[0164] (1) Using the genomic DNA of wild-type Arabidopsis as a template, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com