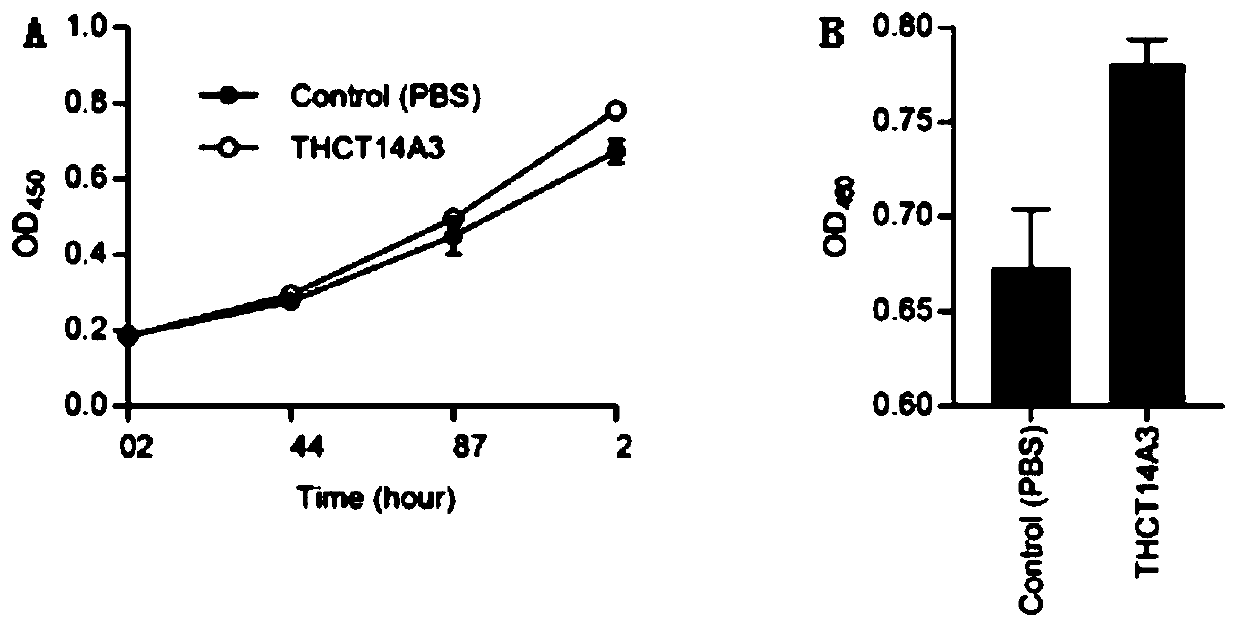
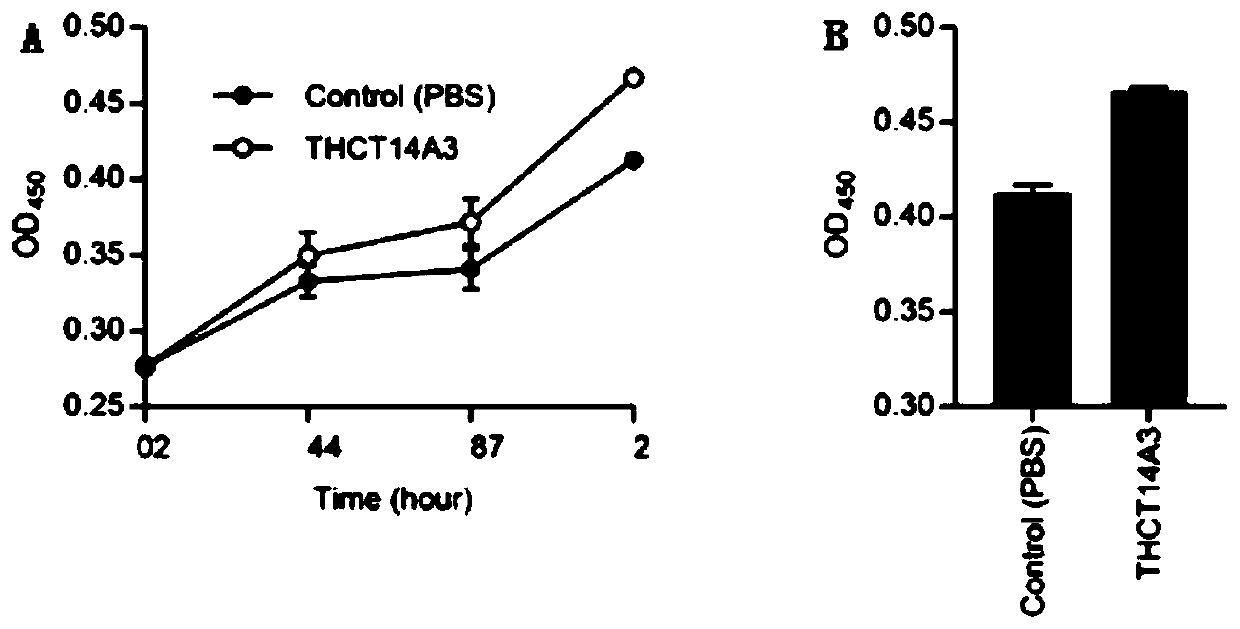
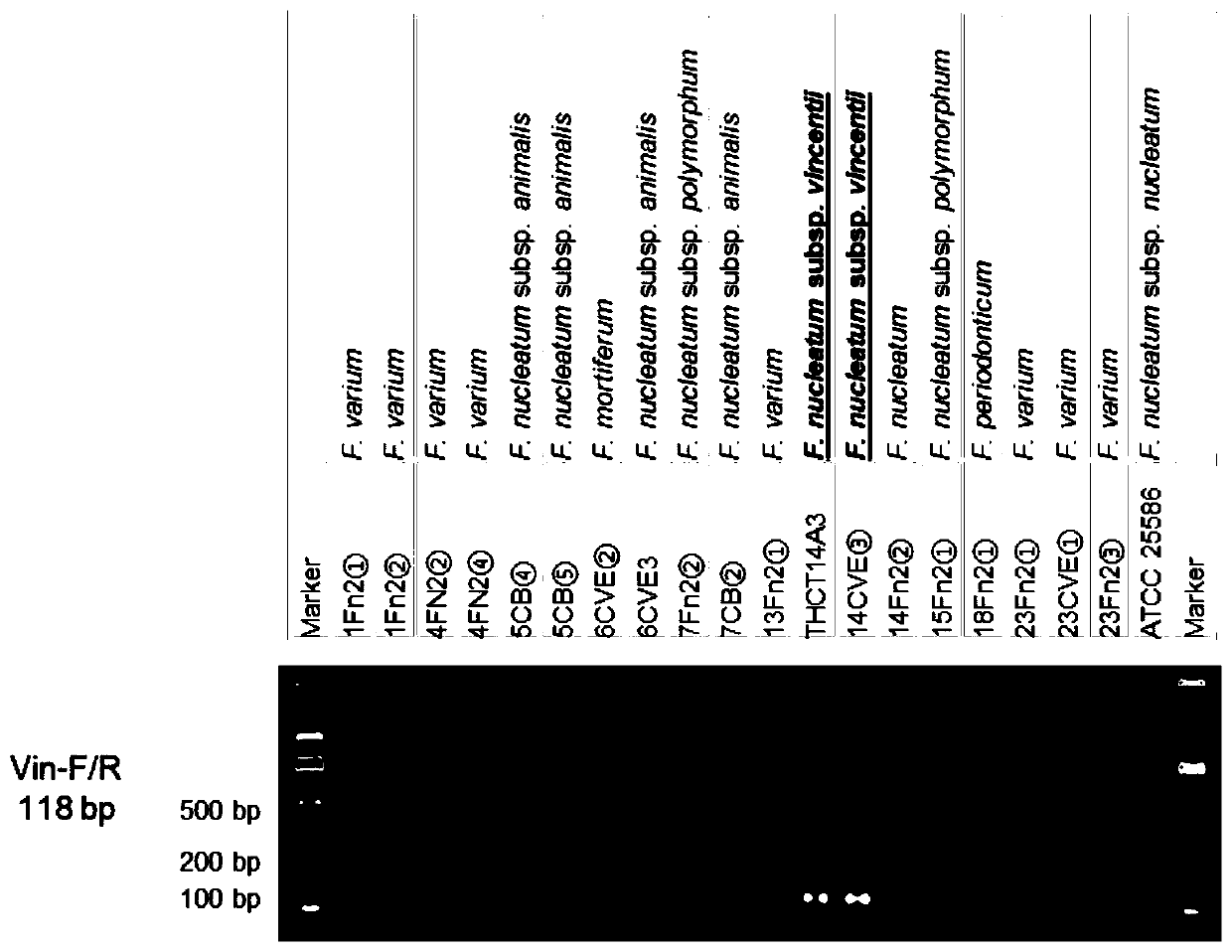
Fusobacterium nucleatum subsp.vincentii strain for intestinal tract separation and application of fusobacterium nucleatum subsp.vincentii strain for intestinal tract separation
A technology of Fusobacterium nucleatum and strains, applied in the direction of bacteria, gastrointestinal cells, and microbial-based methods, which can solve the problems of rare isolates, unclear composition of Fusobacterium nucleatum subspecies, and difficulty in having sufficient representatives of standard strains sexual issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] This example provides a Fusobacterium nucleatum subsp. vincentii strain THCT14A3 isolated from the intestinal tract. The classification of the strain is named Fusobacterium nucleatum subsp.vincentii, and its preservation number is CCTCC NO: M 2019363, and the preservation date is May 2019 On the 17th, the preservation unit was the China Type Culture Collection Center.
[0031] The 16S rRNA gene sequence of the strain is specifically shown in SEQ ID NO: 1, which has 99.59% identity with the 16S rRNA gene sequence (NCBI accession number NR_113040.1) of F. nucleatum subsp. vincentii strain ATCC 51190, and F. nucleatum subsp. The 16S rRNA gene sequence of .nucleatum strain ATCC25586 (NCBI accession number NR_114702.1) has 98.58% identity.
[0032] The complete genome of THCT14A3 was determined, and it was found that the THCT14A3 genome contained a circular chromosome with a size of 2052554bp and a GC content of 27.46%. It contained 1,898 protein-coding genes and 63 non-codi...
Embodiment 2
[0034] In this example, based on the sequence of THCT14A3, combined with comparative genomics, a specific DNA sequence and corresponding primer sequences that can be used to identify Fusobacterium nucleatum subsp. The specific method is as follows:
[0035] On the basis of the THCT14A3 genome, combined with the Fusobacterium nucleatum genome downloaded from NCBI; after subspecies classification according to the ANIb value; through BLASTn analysis of sequences conserved in the genome of subsp. The genomes were compared with BlASTn, in which they were screened for regions not present in the genomes of other subspecies, ie, subspecies Wenckeii-specific DNA sequences (below). (Because of the inevitable polymorphism in the DNA sequence, it is impossible to exhaustively, the DNA characteristic sequence referred to here includes the following sequences and unlisted fragments with BLASTn similarity to any of the following sequences)
[0036] >vincentii_specific_region
[0037] GGCAA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com