Amine dehydrogenase mutant, enzyme preparation, recombinant vector, recombinant cell, preparation methods therefor and application of amine dehydrogenase mutant
A technology of amine dehydrogenase and recombinant cells, which is applied in the biological field and can solve the problems of inability to meet the needs of practical applications and low catalytic efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0055] The method for preparing recombinant cells in the above embodiment includes the following steps S110-S130:
[0056] S110: Perform error-prone PCR amplification on the coding sequence of the wild-type amine dehydrogenase in the above embodiment, and ligate it into an empty vector after digestion to obtain a plasmid of an amine dehydrogenase mutant library.
[0057] Wherein, in the step of performing error-prone PCR amplification on the coding sequence of the wild-type amine dehydrogenase in the above embodiment, the error-prone PCR amplification is carried out by using the primer pair whose sequence is as shown in SEQ ID No.3-SEQ ID No.4. increase. Specifically, the sequence shown in SEQ ID No.3 is: 5'-ACTGCTCATATGGAAAAAATCCGTGTTATCATC-3'; the sequence shown in SEQ ID No.4 is: 5'-TCA GCTCTCGAGTTAAGCGTTGTTAACACCG-3'.
[0058] Among them, the error-prone PCR reaction system is: 0.05U / μL DreamTaq TM , 250 μM of dATP, 250 μM of dGTP, 1050 μM of dCTP, 1050 μM of dTTP, 0.4 μ...
Embodiment 1
[0073] Cloning of the coding sequence of wild-type amine dehydrogenase
[0074] The coding sequence of wild-type amine dehydrogenase (GenBank: CP002131.1) was codon-optimized using Escherichia coli as the host, and was synthesized by Suzhou Jinweizhi Co., Ltd. The sequence used is shown in SEQ ID No.5~SEQ ID No.6 The primer pair used for PCR amplification of the coding sequence of wild-type amine dehydrogenase, PCR amplification using Toyobo (Shanghai) Biotechnology Co., Ltd. KOD high-fidelity polymerase, the amplification conditions are: 95 ° C, 2 min; then 56 ° C, 20sec, 72°C, 90sec, a total of 30 cycles; the last 72°C, 10min. Wherein, the amino acid sequence of the wild-type amine dehydrogenase is shown in SEQ ID No.1; the nucleotide sequence of the wild-type amine dehydrogenase is shown in SEQ ID No.2.
[0075] After the reaction, the PCR product was detected by agarose gel electrophoresis with a mass percent content of 1.5%, and a 1.0 kb band was obtained, the length of ...
Embodiment 2
[0077] Expression, purification and activity determination of amine dehydrogenase
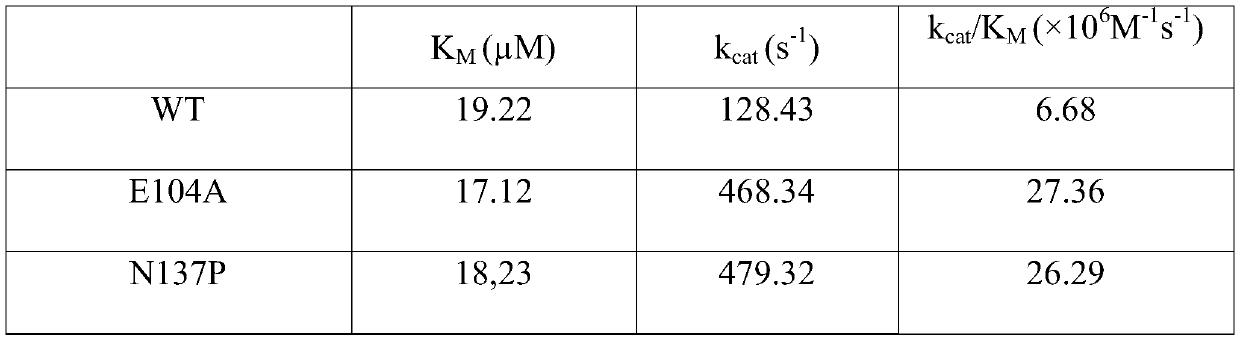
[0078] The engineered bacteria containing the pET28a-ANDD-TDO recombinant plasmid in the glycerol tube were inoculated at 1% by volume into a 4 mL LB medium test tube containing 100 μg / mL kanamycin, and cultured at 37°C and 220rpm for 12h. Transfer all the cultured bacterial liquid to 1L LB medium shake flask containing 50 μg / mL kanamycin, culture at 37°C and 220rpm for about 2.5h, and make the OD 600 When it reached about 0.9, 0.1 mM IPTG inducer was added, and cultured at 25° C. and 200 rpm for 16 hours. The Escherichia coli suspension harvested after induction is ultrasonically disrupted, and then subjected to one-step Ni-NTA affinity chromatography to obtain wild-type amine dehydrogenase with a purity of more than 95%. The activity of wild-type amine dehydrogenase was determined, and the result was: K of wild-type amine dehydrogenase M A value of 19.22 μM, the K of wild-type amine dehydro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com