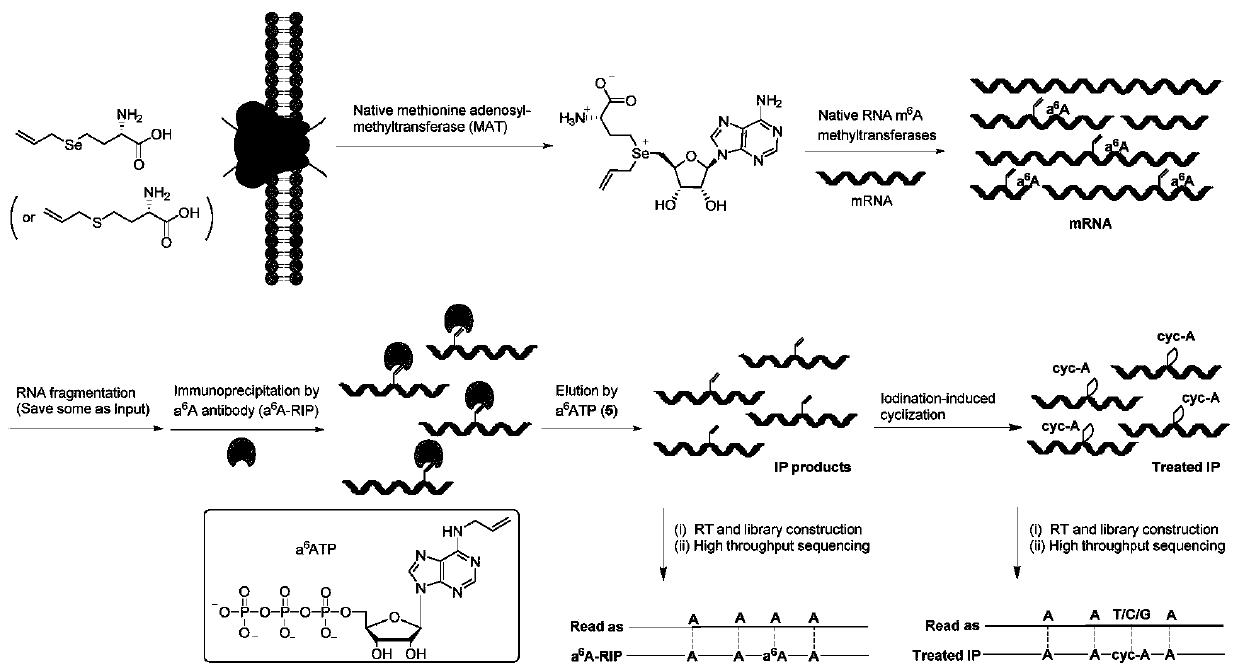
Method and kit for detecting RNA N6-methyladenosine modification at single-base resolution in range of whole transcriptome
A technology of methyl adenine and allyl adenine, applied in biological testing, biochemical equipment and methods, analytical materials, etc., can solve problems such as inability to introduce cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1 In HeLa cells, m on the transcriptome mRNA 6 A detection of modification sites
[0081] 1. HeLa cell culture, allyl labeling and mRNA extraction
[0082] (1) After the HeLa cells were cultured to 80% fullness under normal culture conditions, the medium was aspirated, and the residual medium was washed with phosphate-buffered saline (PBS);
[0083] (2) 10% fetal bovine serum (FBS), 1% 100x penicillin-streptomycin double antibody, and 1 mM cysteine were added to the medium not containing methionine, and the above-mentioned HeLa cells were Incubate in this medium for 30 minutes to remove residual methionine in the cells;
[0084] (3) After removing methionine, add 1 mM allyl-L-seleno / thiohomocysteine to the medium for culturing the above HeLa cells, and continue culturing for 16 hours;
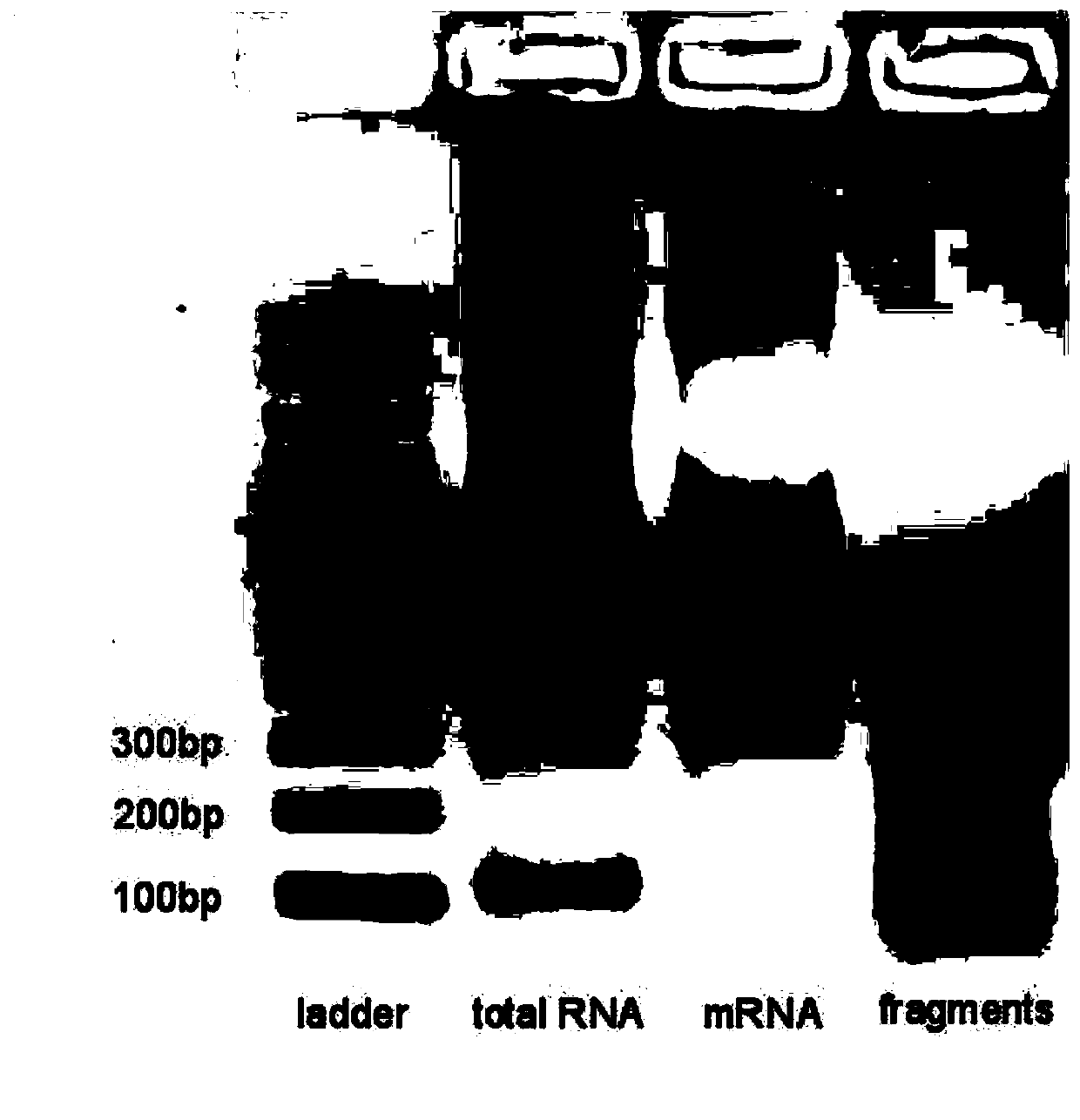
[0085] (4) After the cells have been cultured, suck off the medium, wash off the residual medium with PBS, and then add TRIzol to the culture dish TM Cover the bottom of t...
Embodiment 2
[0164] Example 2 In mouse H2.35 cells, m 6 A detection of modification sites
[0165] 1. Mouse H2.35 cell culture, allyl labeling and mRNA extraction
[0166] (1) Mouse H2.35 cells were cultured under normal culture conditions (with the addition of 200nM DEX, dexamethasone), cultured to 60% of the fullness, sucked off the medium, and washed with phosphate-buffered saline (PBS) Remove residual medium;
[0167] (2) Add 10% fetal bovine serum (FBS), 1% 100x penicillin-streptomycin double antibody, 200nM DEX (dexamethasone), and 1mM cysteine to the medium without methionine , culturing the above-mentioned mouse H2.35 cells in this medium for 30 minutes to remove residual methionine in the cells;
[0168] (3) After removing methionine, add 1 mM allyl-L-seleno / thiohomocysteine to the medium for culturing the above-mentioned mouse H2.35 cells, and continue culturing for 16 hours;
[0169] (4)-(12) The culture, allyl labeling and mRNA extraction of HeLa cells are the same as t...
Embodiment 3
[0175] Example 3 Culture of HeLa cells and mouse H2.35 cells under normal cell culture conditions
[0176] 1. Culture of HeLa cells
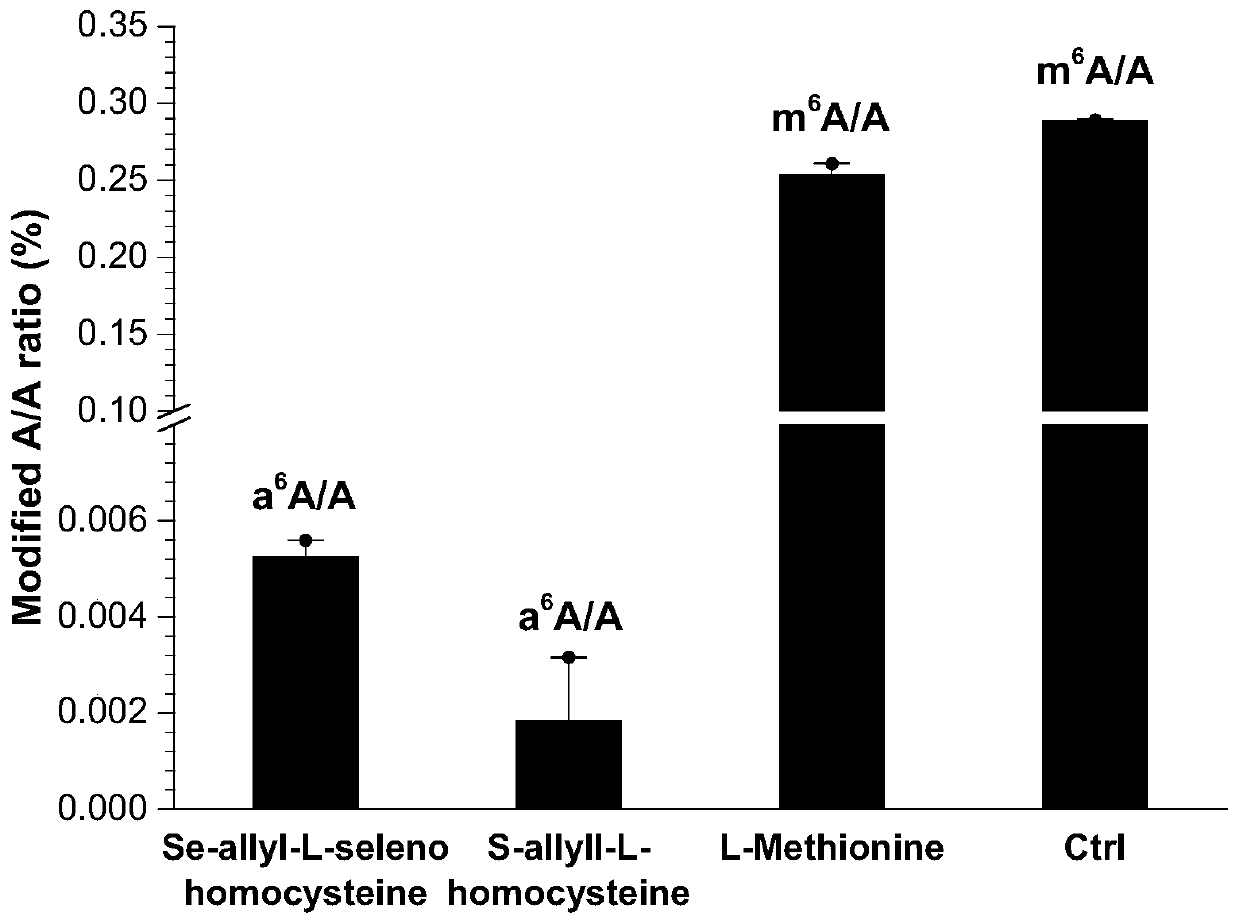
[0177] Add 10% fetal bovine serum (FBS) and 1% 100x penicillin-streptomycin double antibody to normal cell culture medium, culture HeLa cells in this medium for 16-24 hours, then extract HeLa cells mRNA, its m 6 A content such as figure 2 Shown in Ctrl.
[0178] At the same time, HeLa cells were cultured according to the treatment conditions of the above-mentioned allyl labeling method, and 1 mM allyl-L-seleno, allyl-L-thiohomocysteine, or methionine (methionine) was added. Base-L-thiohomocysteine), cultivated for 16 hours, to obtain the m of mRNA 6 A content such as figure 2 In Se-allyl-L-selenohomocysteine, S-allyl-L-homocysteine, L-Methionine shown. figure 2 Se-allyl-L-selenohomocysteine / S-allyl-L-homocysteine is allyl-L-selenohomocysteine / thiohomocysteine introduced into HeLa cells to obtain N in mRNA 6 -The content of allyl ad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com