Drug resistance detection method for vibrio parahaemolyticus fluoroquinolone medicines
A vibrio fluoroquinolone and detection method technology, applied in the field of pathogenic bacteria detection, can solve the problems of inability to detect hidden drug resistance of vibrio parahaemolyticus, cumbersome separation and purification of vibrio parahaemolyticus, unfavorable drug selection and treatment, etc., and achieve reduction Less preparation time, fewer reagents, and less time-consuming effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
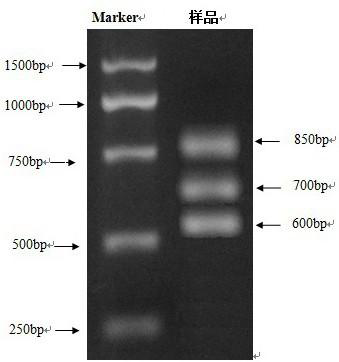
[0019] (1) Cultivate and proliferate Vibrio parahaemolyticus in shake flasks of LB medium. The culture conditions of the shake flasks are 30°C, 18h, 120rpm. After the shake flask culture is completed, 1.0g of live Vibrio parahaemolyticus cells are collected by centrifugation, and weighed with 8mL deionized water. Suspended, repeatedly freeze-thawed 5 times to break the cells, centrifuged at 12000pm for 10min, collected the supernatant, and obtained the template;
[0020] (2) Combine the 5uL template with fluoroquinolone drug resistance gene detection primers GyrA-F, GyrA-R, GyrB-F, GyrB-R, ParE-F and ParE-R (primer concentrations are 25mmol / L, respectively, with 1mM Tris-HCl--0.1mM EDTA buffer dilution) mixed together, then PCR amplification, the PCR amplification reaction system includes ultrapure water, PCR buffer, dNTP at a final concentration of 0.4mmol / L, GyrA-F with a final concentration of 0.4mmol / L, GyrA-R with a final concentration of 0.4mmol / L, GyrB-F with a final co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com