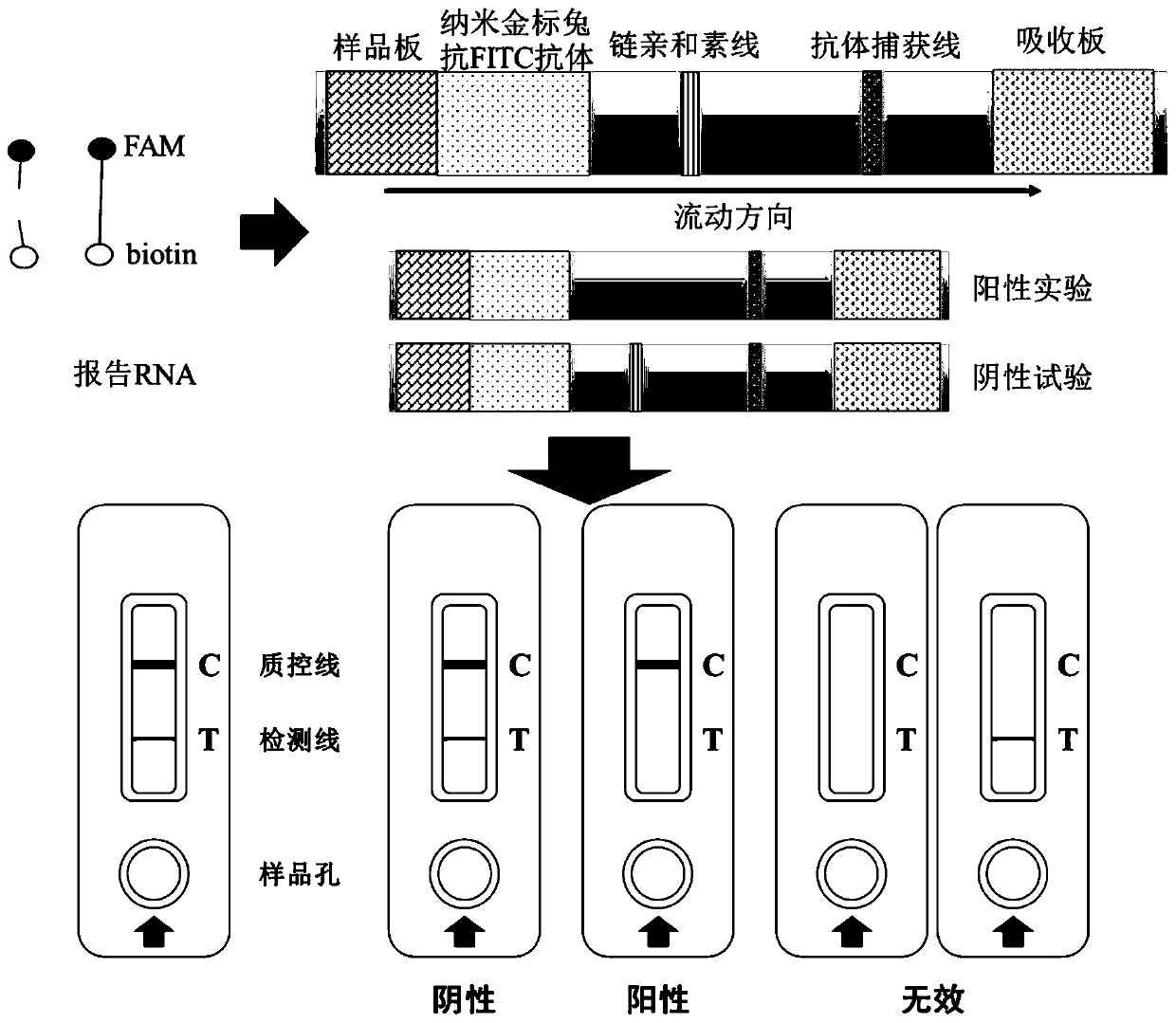
CRISPR nucleic acid detection kit for detecting novel coronavirus (2019-nCoV)
A coronavirus and kit technology, applied in the field of CRISPR nucleic acid detection kits, can solve problems such as insufficient sensitivity, easy contamination, and high requirements for instruments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1. Novel coronavirus nucleic acid detection kit and detection method based on CRISPR-Cas13a system 1. Novel coronavirus nucleic acid detection kit based on CRISPR-Cas13a system
[0065] (1) Preparation of mRNA standard
[0066] The new coronavirus N gene is sequence 3, and the 992-1019th position of sequence 3 is the target sequence of COVID-19 crRNA.
[0067] mRNA is transcribed from the PCR amplification product of the above-mentioned plasmid to simulate viral nucleic acid. The specific method steps are as follows:
[0068] (1) PCR amplification
[0069] Using the synthetic DNA molecule shown in Sequence 3 as a template, PCR amplification was carried out with the system shown in Table 1 below to obtain a PCR product.
[0070] The PCR reaction system was prepared as shown in Table 1.
[0071] Table 1. PCR amplification system
[0072] name volume DNA molecule shown in sequence 3 2μL nCoVnp-F1 2μL nCoVnp-R1 2μL ExTaq Mix...
Embodiment 2
[0139] Example 2. Condition optimization and exploration of detection method based on CRISPR-Cas13a system
[0140] 1. Screening of optimal RT-RAA amplification primers
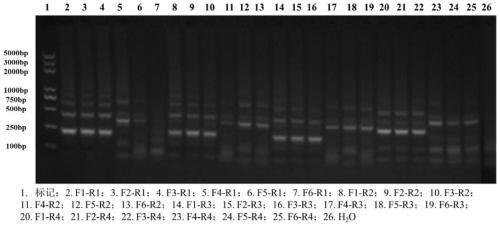
[0141] Taking the mRNA standard product prepared in one (one) in Example 1 as a template, RT-RAA amplification was carried out with the method in (1) of Example 1, and the primers were each primer combination shown in Table 3 ( Combinations such as figure 2 shown) to obtain RT-RAA amplification products.
[0142] The amplification product with water as template was used as a negative control.
[0143] Take 20 μL of the amplified product, add 20 μL of chloroform: Tris equilibrium phenol = 1:1 and mix an equal amount of the reaction product, shake and centrifuge for 10 minutes, discard the supernatant mixture, shake and mix, and centrifuge at 10,000 rpm for 10 minutes , Aspirate 10 μL of supernatant, add 3 μL of 6*Loading Buffer, mix well and run on agarose gel.
[0144] detect as figure 2As shown, the r...
Embodiment 3
[0155] Embodiment 3. Sensitivity detection of the detection method of novel coronavirus nucleic acid based on CRISPR-Cas13a system
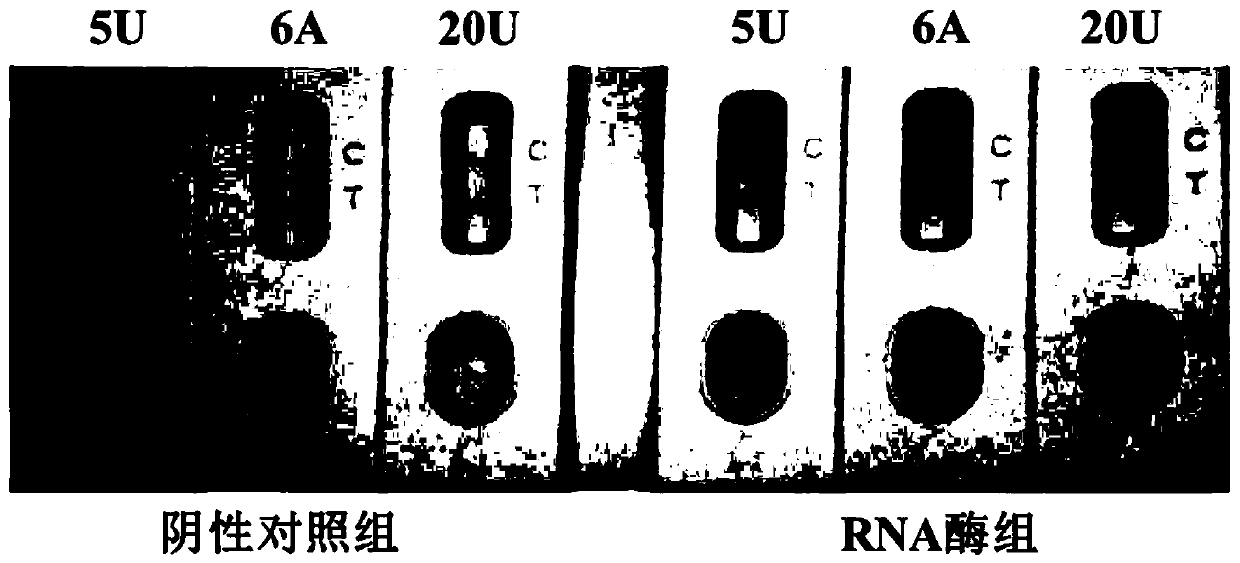
[0156] The mRNA standard substance prepared in one (one) of embodiment 1 utilizes RNase-free water to carry out 10-fold serial gradient dilution, obtains the solution that contains different concentrations of novel coronavirus gene mRNA, detects according to the method for two in embodiment 1, The RT-RAA products in Table 7 were replaced with solutions containing different concentrations of novel coronavirus gene mRNA, so that the concentration of mRNA in the reaction system was 10 5 -10 -1 copies / test, and set the amplification product of water as template as negative control.
[0157] Test results such as Figure 4 shown, 10 5 -10 1 The "T" line disappears and the "C" line appears in the copies / test group; 10 0 copies / test group, 10 -1 Both the "T" and "C" lines in the copies / test group and the negative control group were visible. Judgm...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com