A kind of dna nanostructure, electrochemical aptamer biosensor system and its preparation method and application
A technology of biosensors and nanostructures, applied in the direction of material electrochemical variables, scientific instruments, instruments, etc., to achieve the effect of increasing electrochemical surface area, improving signal amplification ability, and simplifying the experimental process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
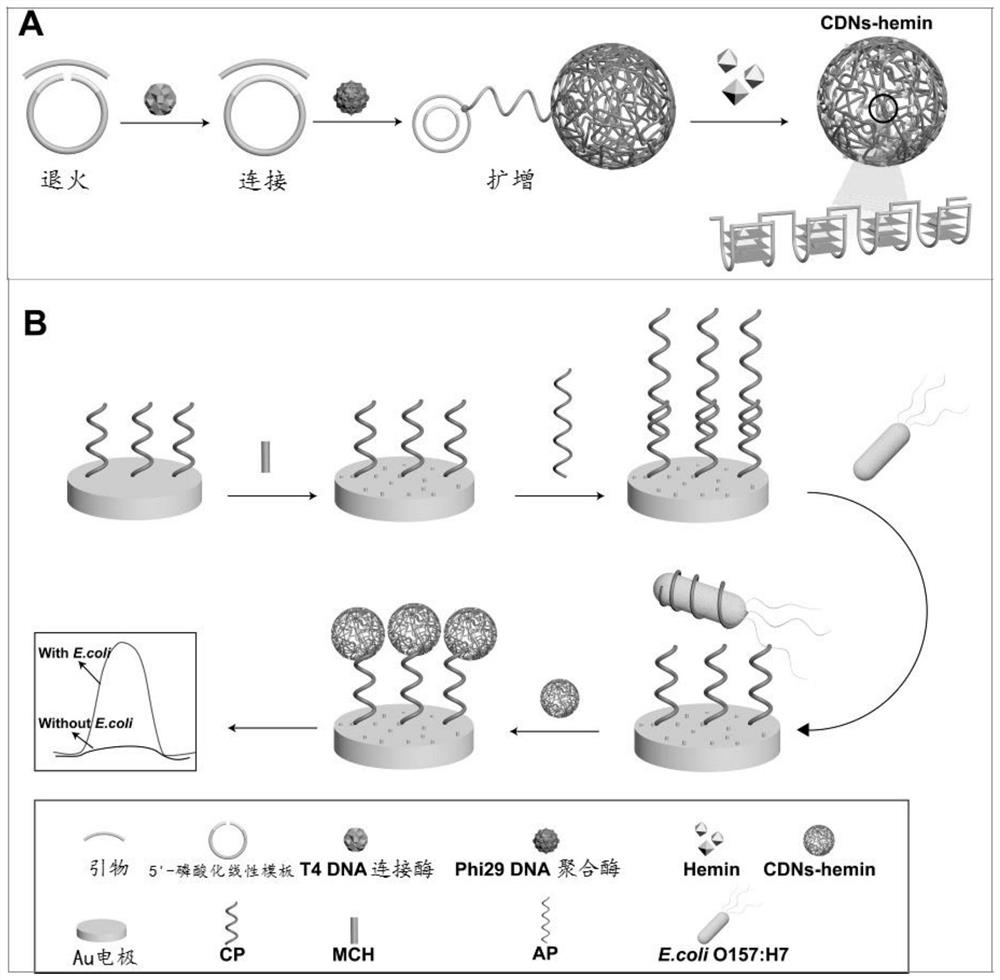
[0066] A kind of preparation method of DNA nanostructure of the present invention, mainly is to synthesize DNA nanostructure (RCA NCs) by rolling circle amplification reaction, comprises the following steps:
[0067] Step 1. Annealing
[0068] In T4 DNA ligase buffer (40mM Tris-HCl, 10mM MgCl 2 , 10mM DTT, 0.5mM ATP, pH7.8), mix the 5'-phosphorylated linear template with the primer, heat the mixture and then cool it to room temperature to form a circular template-primer hybrid;
[0069] Wherein, the nucleic acid sequence of the 5'-phosphorylated linear template is 5'-(PO 4 3- )-ACCCGCCCTACCCAAAATATGCCCTCGGTTGTGGTATTATACGGCATTCCTCTCCGGACAACCCTCGCGGCATCTCGTCCACACTGCCTAAATTTTCCCA-3';
[0070] Wherein, the nucleic acid sequence of the primer is 5'-TTTTGGGTAGGGCGGGTTGGGAAAA-3';
[0071] Step 2. Connect
[0072] Mix the circular template-primer hybrid with T4 DNA ligase and incubate to ligate the circular template gap;
[0073] Step 3. Enzyme inactivation
[0074] After heati...
Embodiment 1
[0111] Synthesis and preparation of embodiment 1 DNA nanostructure
[0112] (1) Annealing
[0113] In T4 DNA ligase buffer (40mM Tris-HCl, 10mM MgCl 2 , 10mM DTT, 0.5mM ATP, pH7.8), mixed 0.3μM 5'-phosphorylated linear template with 0.6μM primer, heated the mixture at 90°C for 5min, then gradually cooled to room temperature within 3h, to form a circular template-primer hybrid;
[0114] The nucleic acid sequence of the 5'-phosphorylated linear template is 5'-(PO 4 3- )-ACCCGCCCTACCCAAAATATGCCCTCGGTTGTGGTATTATACGGCATTCCTCTCCGGACAACCCTCGCGGCATCTCGTCCACACTGCCTAAATTTTCCCA-3';
[0115] The nucleic acid sequence of the primer is 5'-TTTTGGGTAGGGCGGGTTGGGAAAA-3'.
[0116] (2) connection
[0117] Mix the circular template-primer hybrid with 10 U of T4 DNA ligase and incubate at 16°C for 16 hours to ligate the circular template gap.
[0118] (3) Enzyme inactivation
[0119] Heat at 65°C for 10 min to inactivate T4 DNA ligase.
[0120] (4) Amplification
[0121] Add the 2mM clos...
Embodiment 2
[0126] Hemin / G-quadruplex DNAzyme functionalization of Example 2RCA NCs
[0127] (1) Treat 25 μL of TE buffer containing RCA NCs at 90°C for 1 min, cool to 4°C, and keep for 60 min.
[0128] (2) Add the product obtained in step (1) together with 100 μM Hemin (heme) solution to 50 μL Tris-HCl buffer, and incubate at 37°C for 60 min, so that Hemin (heme) can be embedded in the RCA NCs In the G-quadruplex unit, functionalized RCA NCs were formed, that is, functionalized Hemin / G-quadruplex DNAzyme.
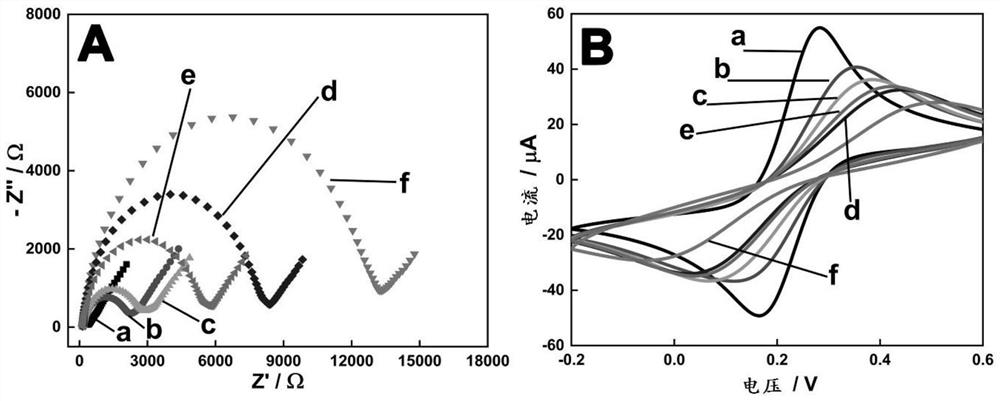
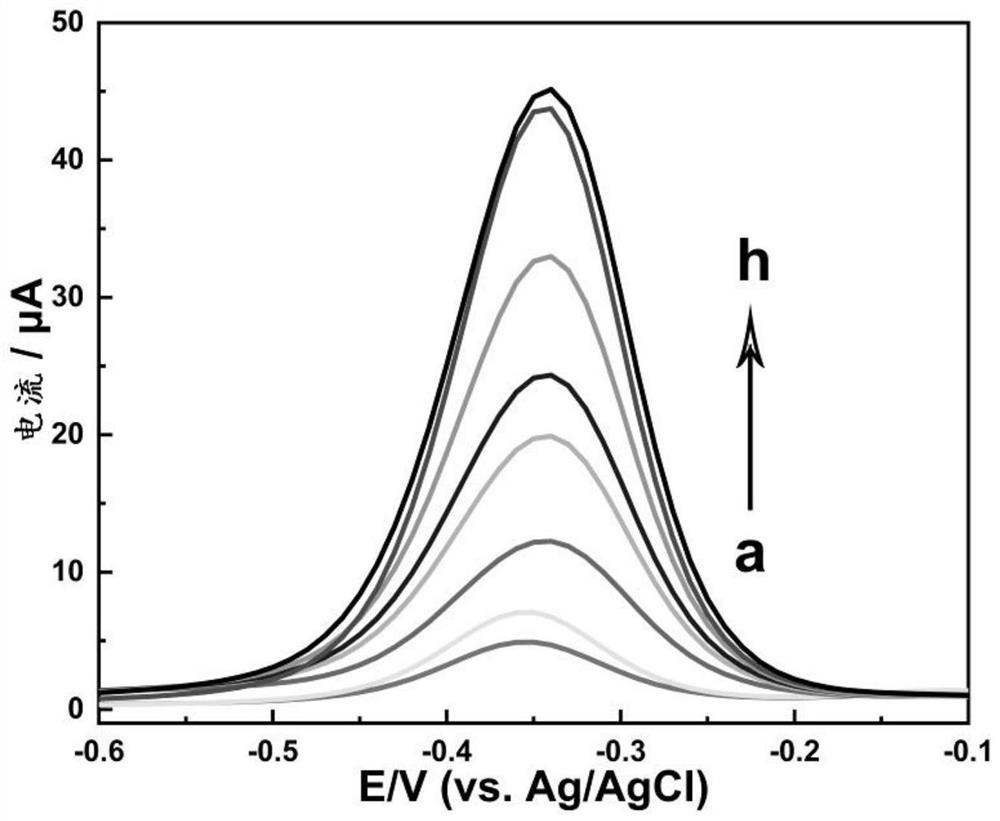
[0129] (3) Store the prepared functionalized Hemin / G-quadruplex DNAzyme below 4°C. RCA NCs with peroxidase-mimicking DNAzyme activity can catalyze the substrate H 2 o 2 reduction, resulting in an electrochemical signal.
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com