Method for detecting viable count in pre-coated feed microecological preparation product
A technology of micro-ecological preparations and detection methods, applied in the preparation of test samples, measurement devices, color/spectral characteristics measurement, etc., can solve the problems of inapplicability, no indication of adenosine, and no consideration of bacterial cell damage, etc. , to achieve the effect of small influence of human factors, short detection cycle and good reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
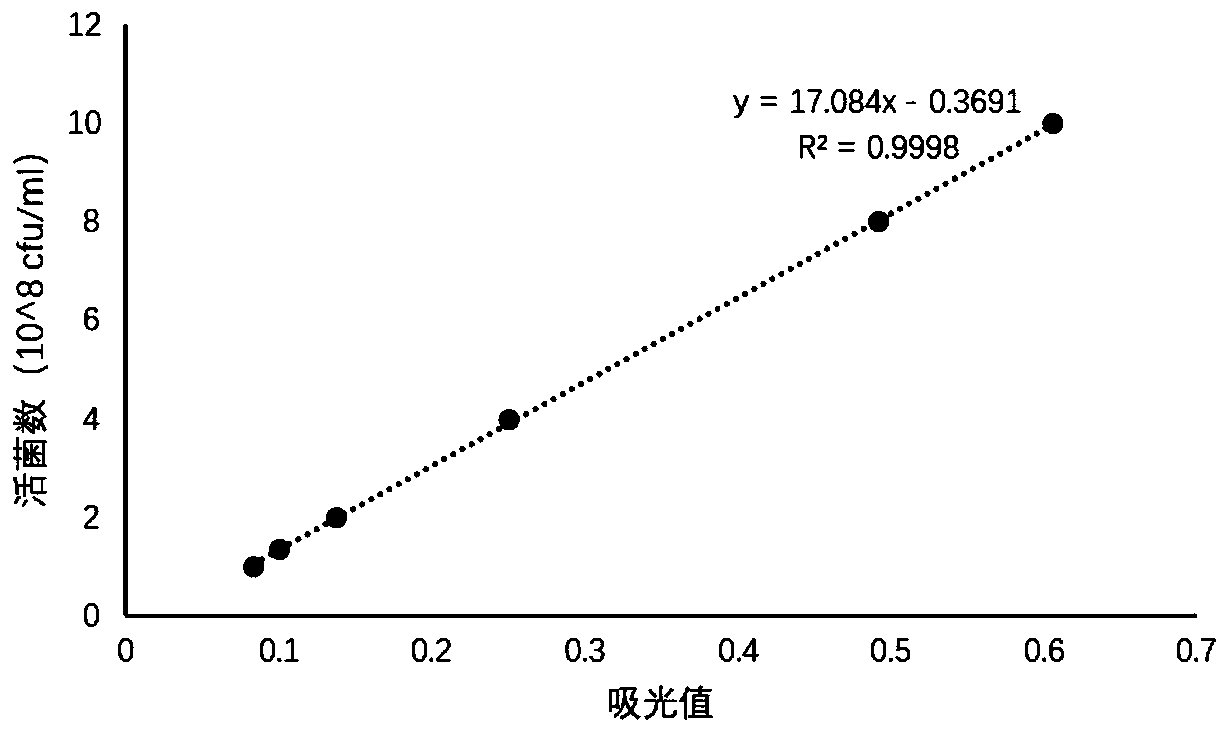
[0044] In this embodiment, taking lactic acid bacteria as an example, a method for detecting the number of viable bacteria in a pre-coated microecological preparation product for feeding according to an embodiment of the present invention is described in detail. The principles and methods of other types of probiotics are the same as those of the examples here.
[0045] A method for detecting the number of live bacteria in coated lactic acid bacteria, comprising the following steps (taking the sample of coated lactic acid bacteria before the amount of bacteria is 1×1010 as an example):
[0046] Step 1: Draw the standard curve
[0047] (1) Cultivate free lactic acid bacteria for 21 hours, calculate the number of live bacteria by gradient dilution, and take an appropriate amount of bacterial liquid to centrifuge, add an equal volume of sterile water and resuspend in sterile water to obtain a bacterial suspension, and set aside;
[0048] (2) Take the bacterial solution from step ...
Embodiment 2
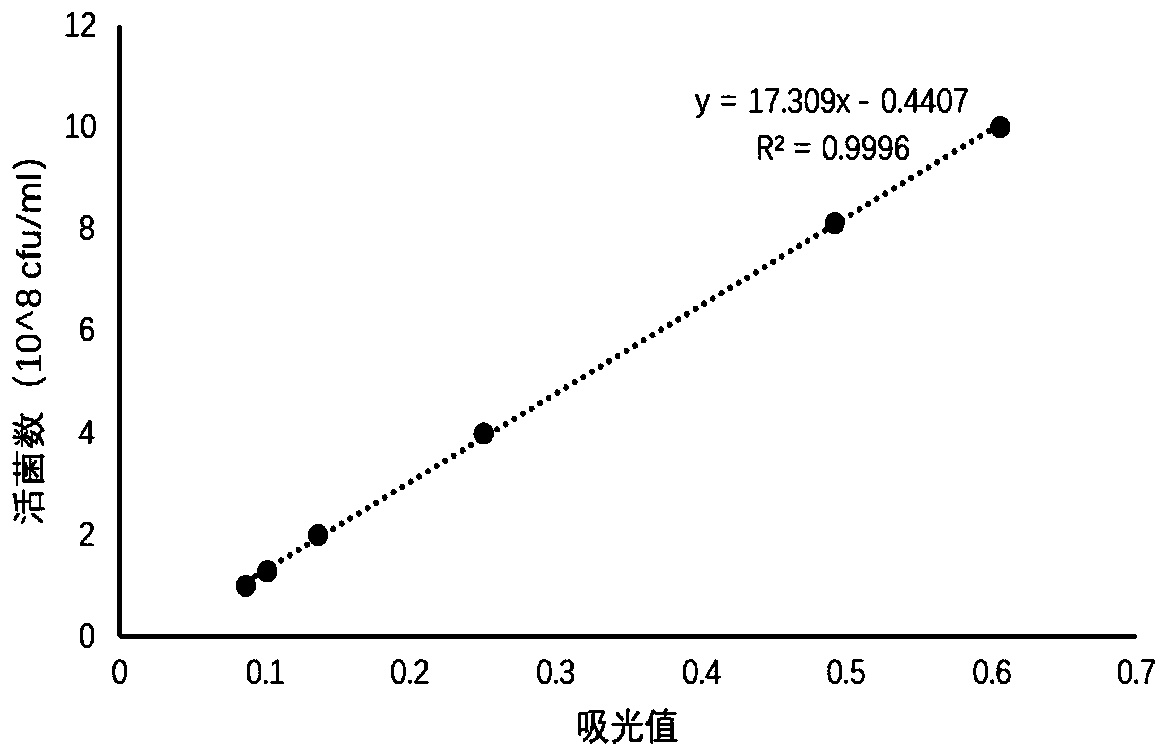
[0067] In this embodiment, taking lactic acid bacteria as an example, a method for detecting the number of viable bacteria in a pre-coated microecological preparation product for feeding according to an embodiment of the present invention is described in detail. The principles and methods of other types of probiotics are the same as those of the examples here.
[0068] A method for detecting the number of live bacteria in pre-coated lactic acid bacteria, comprising the following steps:
[0069] Step 1: Draw the standard curve
[0070] (1) Cultivate free lactic acid bacteria for 23 hours, calculate the number of live bacteria by gradient dilution, and take an appropriate amount of bacterial liquid to centrifuge, add an equal volume of sterile water and resuspend to obtain a bacterial suspension, and set aside;
[0071] (2) Take the bacterial solution from step (1), and after diluting different gradients, take 100 μL of the bacterial solution and add it to a 96-well microtiter ...
Embodiment 3
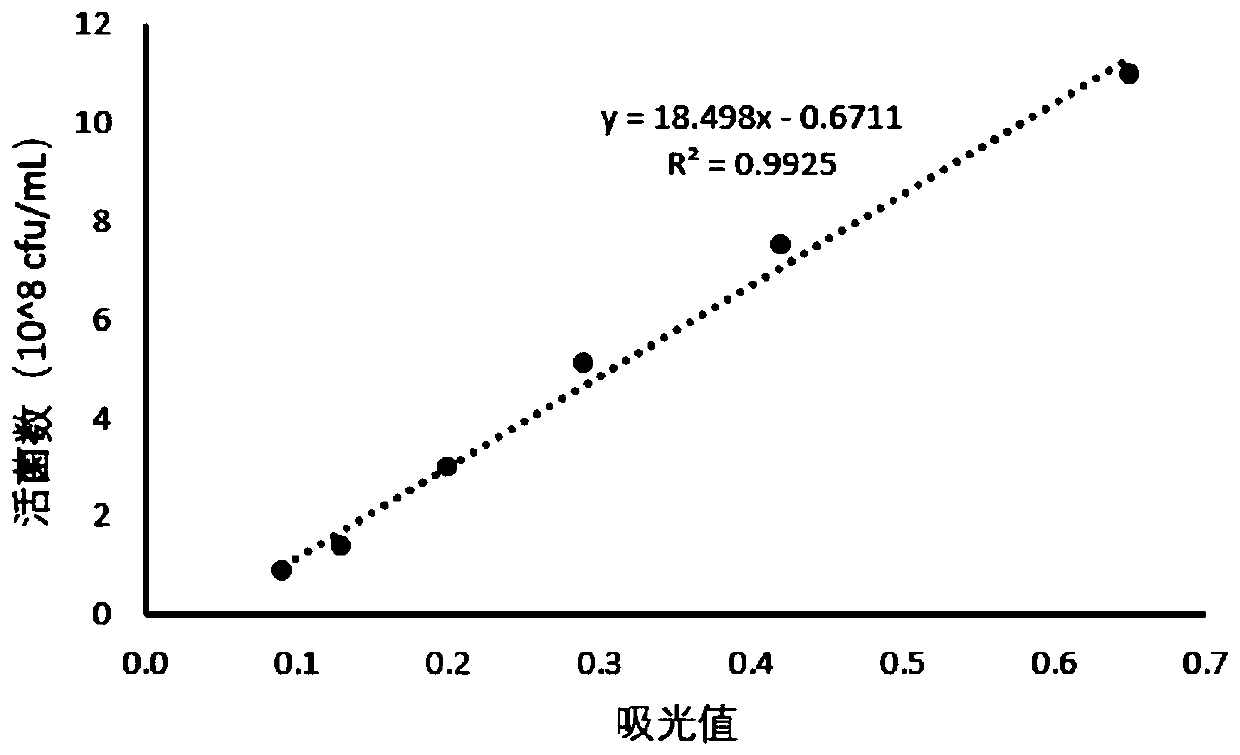
[0090] In this embodiment, taking Saccharomyces cerevisiae as an example, a method for detecting the number of viable bacteria in a pre-coated probiotic product for feeding according to an embodiment of the present invention is described in detail. The principles and methods of other types of probiotics are the same as those of the examples here.
[0091] A method for detecting the number of live bacteria in Saccharomyces cerevisiae, comprising the following steps:
[0092] Step 1: Draw the standard curve
[0093] (1) Cultivate free Saccharomyces cerevisiae for 38 hours, calculate the number of live bacteria by gradient dilution, and take an appropriate amount of bacterial liquid to centrifuge, add an equal volume of sterile water to resuspend to obtain a bacterial suspension, and set aside;
[0094] (2) Take the bacterial solution from step (1), and after diluting different gradients, take 100 μL of the bacterial solution and add it to a 96-well microtiter plate, and add 50 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com