Engineering bacterium for efficiently synthesizing pyruvic acid and D-alanine and construction method and application of engineering bacterium
A technology of engineering bacteria and pyruvate, applied in the field of metabolic engineering, can solve problems such as long catalysis time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Molecular docking and selection of saturation mutation sites
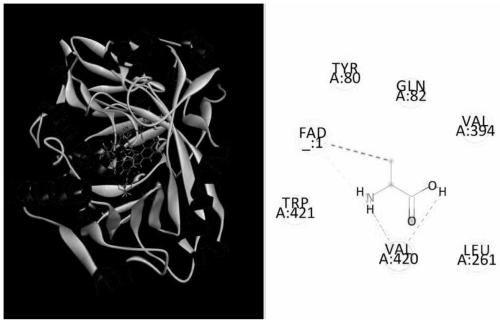
[0038] Search the crystal model of its homologous enzyme in the SWISS MODEL database. Model 5fjm.1A has high homology with pm1, and its homology is 93.51%. Comparing its amino acid series with that of pm1, the results show that only a few regions have different amino acids, which shows that the crystal structure of 5fjm.1A is basically consistent with that of pm1. Therefore, model 5fjm.1A was selected for molecular docking with L-alanine.
[0039] Semi-flexible molecular docking experiments were performed using Autodock software. According to the operation manual of the Autodock software, the three-dimensional protein structure file of the model 5fjm.1A (including the structure of the coenzyme FAD) and the three-dimensional structure file of the ligand L-alanine were molecularly docked using the Lamarck genetic algorithm (LGA).
[0040] Select the characteristic sites of site-directed saturation ...
Embodiment 2
[0041] Example 2: Establishment of Site-Directed Saturation Mutation Library
[0042] Table 1
[0043]
[0044] According to the gene sequence of pm1, design the primers shown in the above table, use the above primers and use the pET20b-pm1 plasmid as a template to amplify the whole plasmid by PCR, transfer the PCR products into E. on the tablet.
Embodiment 3
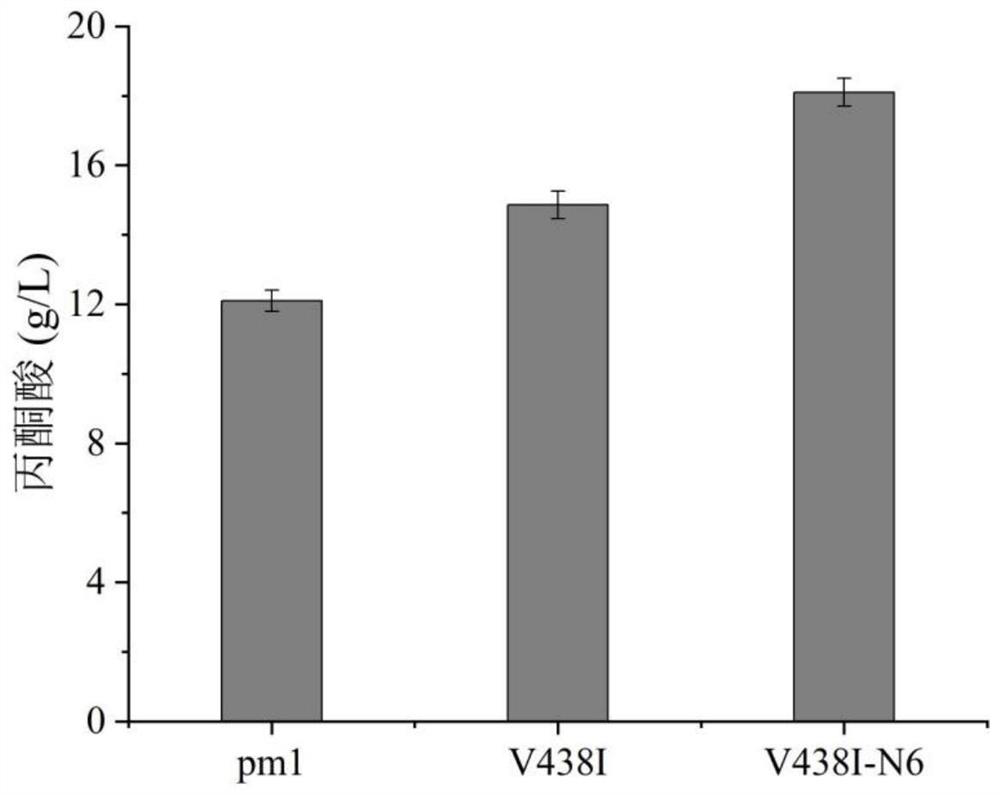
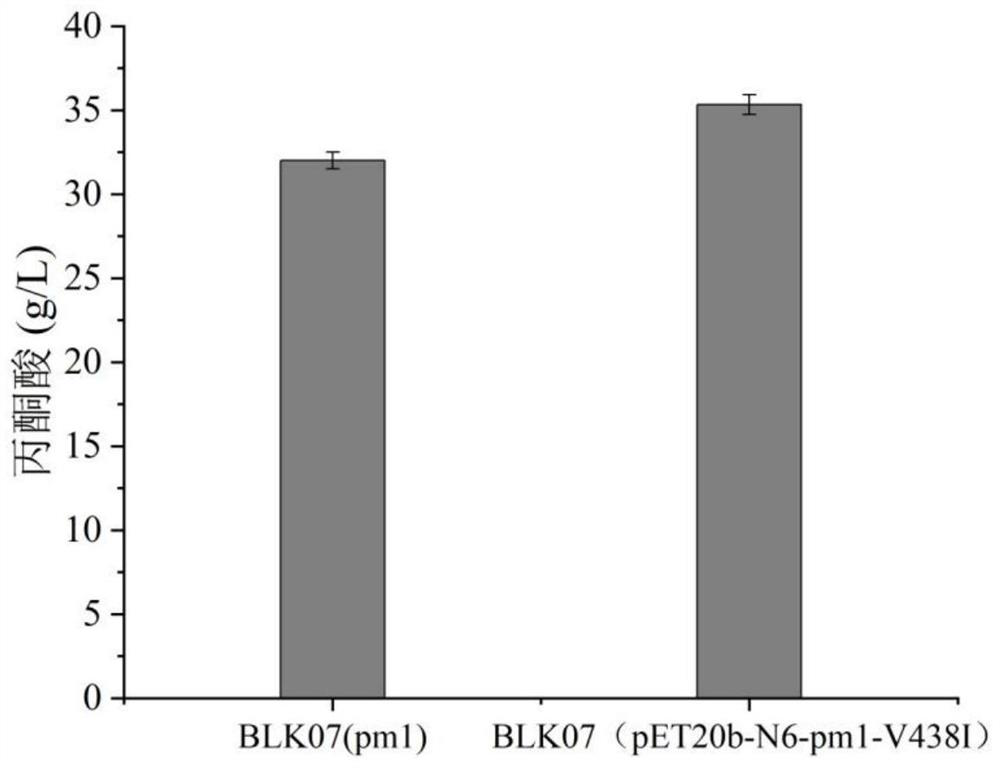
[0045] Example 3: High-throughput screening and validation
[0046] Use a toothpick to pick about 100 single clones from each saturated mutation point library and transfer them to a 96-well plate for seed culture. Each well contained 600 μl of ampicillin-resistant liquid LB medium, and cultured at 37° C. for 12 hours on a well plate shaker. Afterwards, 200 μl of the seed solution was transferred to a 96-well plate containing 600 μl of ampicillin-resistant liquid TB medium per well for fermentation induction culture, and at the same time, IPTG (final concentration 0.01 mM) was added to induce at 37° C. for 6 h and then the fermentation was terminated. Collect the cells by centrifugation (3,500rpm, 5min, 4°C) in the fermentation broth, discard the supernatant and add 200μl 40g / L L-alanine (dissolved in 0.2M pH 7.0 phosphate buffer), and place on a plate shaker at 37°C The reaction was started in 30 min, and the reaction was stopped by centrifugation after 30 min, and the conten...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com