Method for improving mannase activity of bifunctional cellulase, cellulase mutant RMX-M and application
A technology of mannanase and cellulase, applied in the field of agricultural biology, can solve the problems that the mutation scheme is difficult to obtain the expected technical effect, and the research between sequence and function is limited.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Cellulase mutant recombinant vector with altered substrate specificity pPIC9r-RMX-M preparation of
[0032] The wild-type cellulase coding gene before mutation (removing the signal peptide coding sequence) was cloned into the expression vector pPIC-9r, and the recombinant vector was named pPIC9r-RMX ; with recombinant vector pPIC9r-RMX As a template, it is amplified by primers carrying mutation sites to obtain recombinant vectors carrying mutant sequences, named as pPIC9r-RMX-M .
[0033] Table 1 Cellulase mutants with altered substrate specificity RMX-M specific primer
[0034] Primer sequence (5' — 3') a
Embodiment 2
[0035] Example 2 Preparation of cellulase mutants with altered substrate specificity.
[0036] (1) Cellulase mutants RMX-M Mass expression at the shake flask level in Pichia pastoris
[0037] Mutant genes containing altered substrate specificity will be obtained RMX-M recombinant plasmid pPIC9r-RMX-M Transform Pichia pastoris GS115 to obtain recombinant yeast strain GS115 / RMX-M . Take the GS115 strain containing the recombinant plasmid, inoculate it in a 1 L Erlenmeyer flask with 300 mL of BMGY medium, and culture it on a shaker at 220 rpm at 30°C for 48 h; centrifuge the culture solution at 4,000 g for 5 min, discard the supernatant, and use 200 mL for the precipitation. Resuspend in BMMY medium containing 0.5% methanol, and place again at 30°C, 220 rpm to induce culture. 1 mL of methanol was added every 12 h, and the supernatant was taken for enzyme activity detection.
[0038] (2) Purification of recombinant cellulase
[0039] The recombinant cellulase supernata...
Embodiment 3
[0040] Example 3 Activity Analysis of the Cellulase Mutant and Wild Type Improved by Recombinant Mannanase Activity
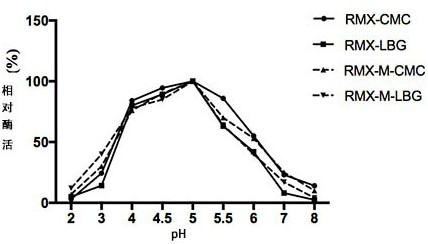
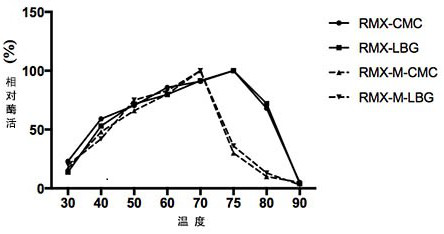
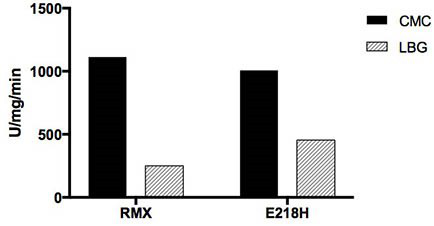
[0041] The basic enzymatic properties of recombinant cellulase RMX and mutant RMX-M were determined by dinitrosalicylic acid (DNS) method. The specific method is as follows: at pH 5.0, 75 °C, 1 mL of reaction system includes 100 µL of appropriate diluted enzyme solution, 900 µL of substrate, react for 10 min, add 1.5 mL of DNS to terminate the reaction; boil in boiling water for 5 min and then cool down To room temperature, measure the OD value at a wavelength of 540 nm. Mannanase assay method is the same as cellulase. Definition of endo-cellulase activity unit: Under certain conditions, the amount of enzyme required to decompose the substrate to generate 1 μmoL glucose per minute is 1 activity unit (U). Definition of mannanase activity unit: Under certain conditions, the amount of enzyme required to decompose a substrate to generate 1 μmoL mannose per minute...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com