A kind of endoxylanase mutant s35f07 and its preparation method and application
An endoxylanase, S35F07 technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, applications, etc., can solve problems such as limited applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Construction of embodiment 1 mutant library
[0032] The construction process of the mutant library is as follows:
[0033] (1) Extract the genomes of Arthrobacter sp. and Lechevalieria sp. according to the instructions of the GENE STAR Bacterial Genome Extraction Kit;
[0034](2) According to the Arthrobacter sp. endoxylanase nucleotide sequence JQ863105 (SEQ ID No.3) recorded in GenBank, primers 5'-GTGCAGCCGGAGGAAAAACG-3'(SEQ ID No.5) and 5'-GATGAAGGCAGGATCCGGGGT-3'(SEQ ID No.6), using the Arthrobacter sp. genome as a template for PCR amplification to obtain the endoxylanase gene xynAGN16L; Lechevalieria sp. endoxylanase nucleotide sequence JF745868 (SEQ ID No.4), designed primers 5'-GTCTCGGCCCCGCCGGACGT-3'(SEQ ID No.7) and 5'-GGCTCGCTTCGCCAGCGTGG-3' (SEQ IDNo.8), using the Lechevalieria sp. genome as a template for PCR amplification to obtain the endoxylanase gene xynAHJ3; the PCR reaction parameters are: denaturation at 94°C for 5min; then 94°C Denaturation for 3...
Embodiment 2
[0040] Screening of embodiment 2 mutants
[0041] The screening process of mutants is as follows:
[0042] (1) Take 2 μL of bacterial liquid from the 96-well cell culture plate of the preserved mutant library, and inoculate it into 200 μL / well liquid LB culture medium (containing 100 μg mL -1 Amp) in a 96-deep-well plate at 37°C with shaking at 200rpm until OD 600 >1.0 (about 20h), add 2mM IPTG and 100μg mL -1 200 μL liquid LB culture solution of Amp, induced overnight at 20°C, 160 rpm;
[0043] (2) Add 40 μL / well of PopCulture after induction TM Cell lysate, shake and lyse the cells at 25°C for 30 minutes;
[0044] (3) Take 50 μL of McIlvaine buffer (pH=7.0) containing 1.0% (w / v) beech xylan and 50 μL of cell lysate, and react in a 96 deep-well plate in a 70° C. incubator for 2 hours. After the reaction, add 150 μL of DNS reagent to terminate the reaction, incubate in a 140°C incubator for more than 20 minutes and cool to room temperature, and use a microplate reader to ...
Embodiment 3
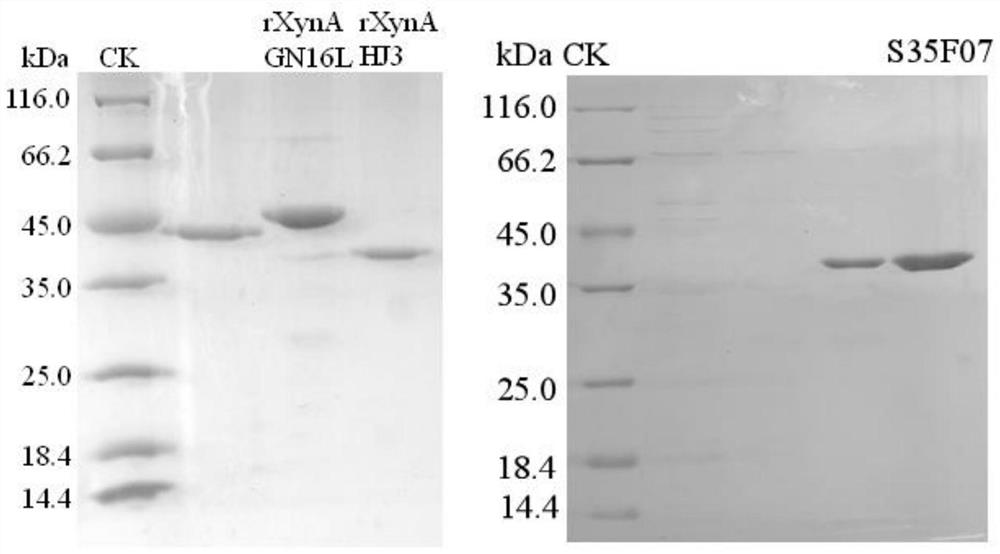
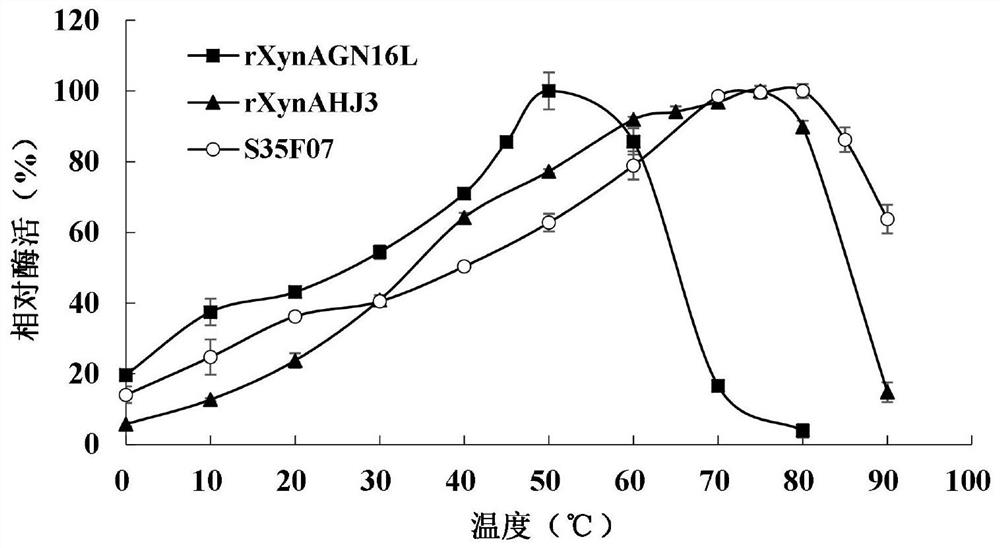
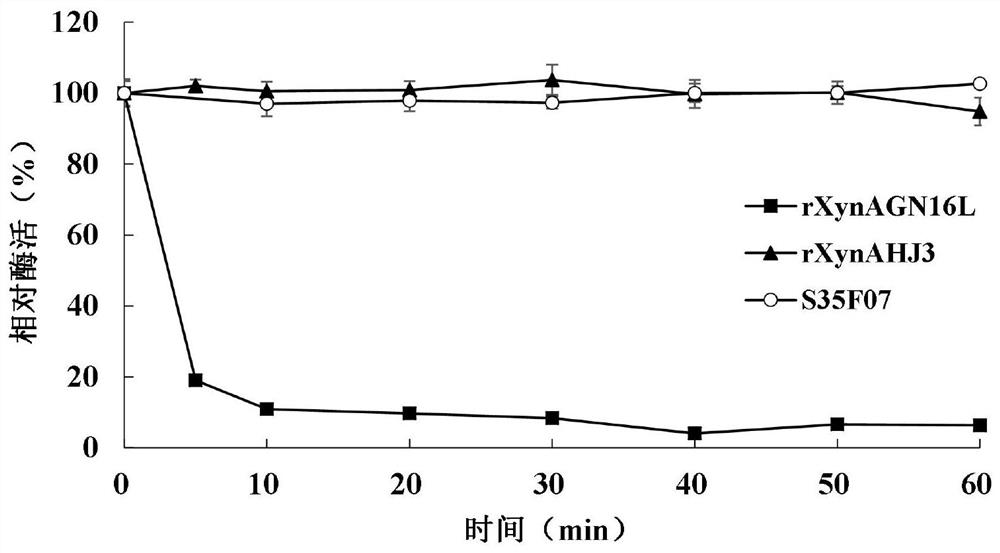
[0048] Example 3 Enzyme Preparation of Mutant S35F07 and Wild Enzymes rXynAGN16L and rXynAHJ3
[0049] The recombinant strains containing mutant S35F07, wild enzymes rXynAGN16L and rXynAHJ3 were inoculated in LB (containing 100 μg mL -1 Amp) medium, shake rapidly at 37°C for 16h.
[0050] Then, the activated bacterial solution was inoculated into fresh LB (containing 100 μg mL -1 Amp) culture medium, rapid shaking culture for about 2–3h (OD 600 After reaching 0.6-1.0), add IPTG (isopropylthiogalactopyranoside) at a final concentration of 0.1 mM for induction, and continue shaking culture at 20° C. for about 20 h. Centrifuge at 12000rpm for 5min to collect the bacteria. After suspending the bacteria with an appropriate amount of pH=7.0 Tris-HCl buffer solution, the bacteria were disrupted ultrasonically in a low-temperature water bath.
[0051] The crude enzyme solution concentrated in the cells above was centrifuged at 13,000rpm for 10min, the supernatant was aspirated, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com