Genetically encoded formaldehyde reactive unnatural amino acid, preparation method and application thereof
A non-natural amino acid, genetic coding technology, applied in the preparation of carbamate derivatives, the preparation of aminohydroxy compounds, the preparation of organic compounds and other directions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
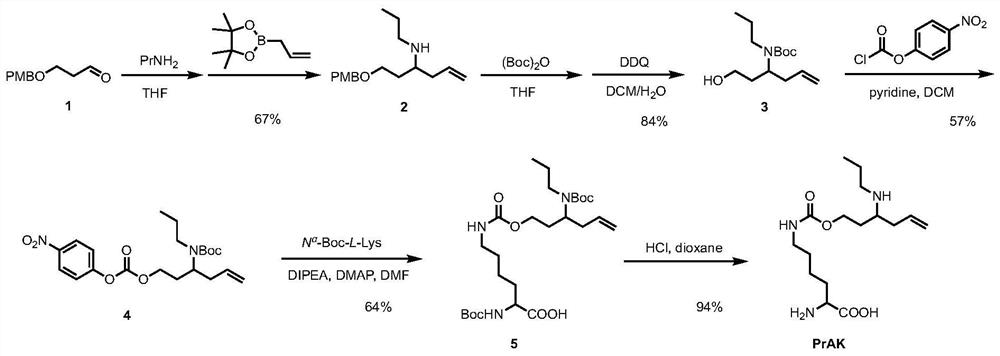
[0115] Example 1 Design and preparation of unnatural amino acid lysine analogue PrAK
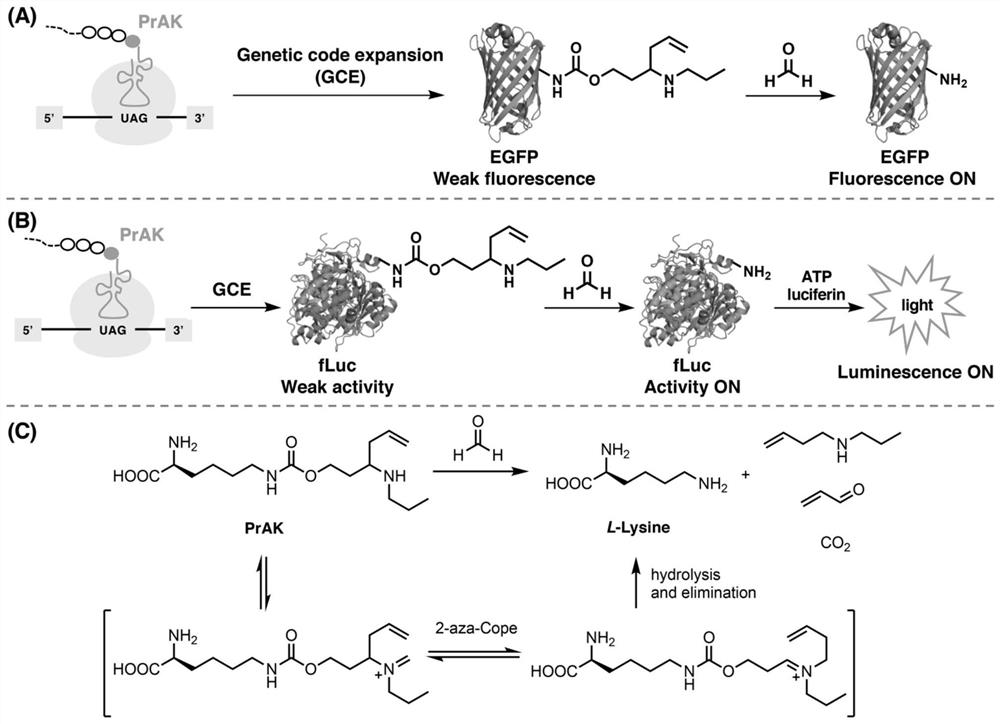
[0116] The original idea of the present invention is to synthesize an unnatural amino acid with formaldehyde reactivity and specifically insert it into biological macromolecule fluorescent and bioluminescent proteins to control their fluorescence and bioluminescence ( figure 1 A, 1B). When formaldehyde is not present, the EGFP and fLuc mutants modified by unnatural amino acids have no fluorescence and bioluminescence activity; when formaldehyde is present, the unnatural amino acid reacts with formaldehyde to recover to key lysine residues, resulting in fluorescence and bioluminescence. Luminous activity recovery ( figure 1 A, 1B), so as to realize formaldehyde detection. In order to achieve the above objective, the present invention independently designs a formaldehyde-reactive lysine analogue PrAK, which is proved by the detection and imaging of formaldehyde in biological samples.
[0117] Th...
Embodiment 2
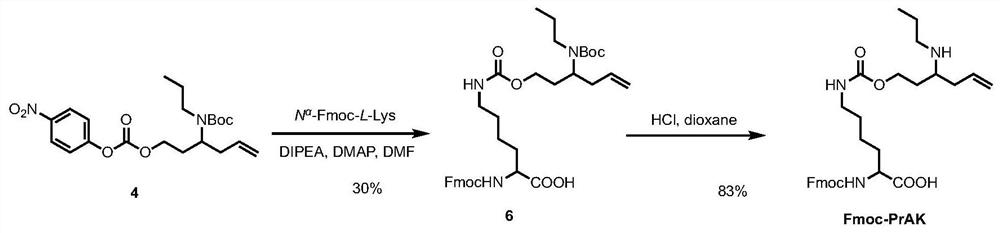
[0129] Example 2 In vitro Fmoc-PrAK and formaldehyde reactivity experiment
[0130] 1) Prepare Fmoc-PrAK, the specific method steps are as follows (see image 3 ):
[0131] Compound 6 Synthesis.
[0132] To a solution of Na-Fmoc-L-Lys (190 mg, 0.474 mmol) in DMF (5 mL) was added DIPEA (100 μL, 0.61 mmol) and compound 4 (143 mg, 0.34 mmol) in DMF (5 mL). The reaction mixture was stirred at 25°C for 10 h and quenched with water. Separate each layer, use CH for water layer 2 Cl 2 (50 mL) extraction. The combined organic layer was washed with 1N HCl (3×15 mL) and brine (20 mL), and washed with anhydrous Na 2 SO 4 Dry and concentrate under reduced pressure. The residue was purified by silica gel column chromatography (5% MeOH / CH 2 Cl 2 ), a white solid compound 6 (86 mg, 30% yield) was obtained. 1 H NMR(300MHz,d 6 -DMSO)δ7.89(d,J=7.3Hz,2H), 7.73(d,J=7.1Hz,2H), 7.60(d,J=7.8Hz,1H), 7.40(dd,J=14.1,6.8 Hz, 2H), 7.33 (dd, J = 14.1, 6.8 Hz, 2H), 7.09 (s, 1H), 5.81-5.57 (m, 1H), 5.05-5.00 (...
Embodiment 3
[0138] Example 3 Site-specific introduction of unnatural amino acid PrAK into protein
[0139] 1) PrAKRS sequence optimization and screening
[0140] In order to obtain the active site mutant of pyrrolysyl tRNA synthetase capable of inserting PrAK, the present invention modeled PrAK computational simulation into the pyrrolysine binding pocket of pyrrolysyl tRNA synthetase, and noted the PrAK The side chain may conflict in space with many residues around the pocket (for example, Y306, L309, C348, M350, I405, and I413V). Therefore, a series of active site mutants based on wild-type pyrrolysyl tRNA synthetase were constructed by mutating these residues into amino acids with less steric hindrance through rational design, as shown in the table below.
[0141] Mutation group Mutation site Group 1 L309A,Y384F Group 2 L309A,C348S,Y384F Group 3 Y384F Group 4 Y306A,Y384F Group 5 Y306G, L309G, Y384F Group 6 L309A,Y348F,Y384F Group 7 Y306G, L309A, Y384F Group 8 Y306A, L309G, Y384F ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com