Label-free nuclease analysis method based on stable isotope detection
A stable isotope and analysis method technology, applied in the field of label-free nuclease analysis based on stable isotope detection, can solve the problems of low stability of CuNPs, long-term detection and real-time monitoring, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 The label-free analysis method based on stable isotope detection to exonuclease (ExoI) detection
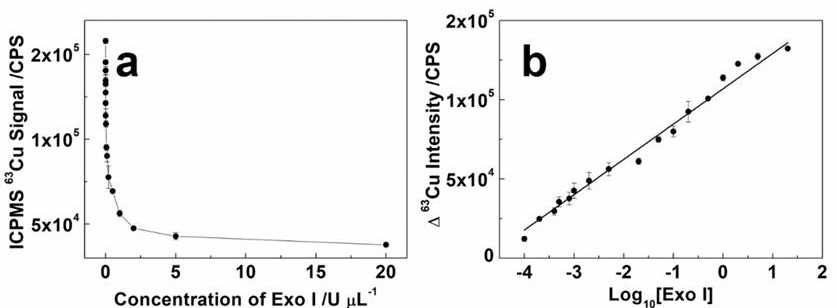
[0043] exonuclease (ExoI) is a nuclease that can specifically cut single-stranded DNA (ssDNA) from the 3' end to the 5' end. Meanwhile, single-stranded DNA (PolyT-ssDNA) composed entirely of thymines can be used as a DNA template to synthesize CuNPs natively. Therefore, stable isotope-based label-free detection of ExoI can be performed using PolyT-ssDNA.
[0044] In the experiment, the concentration of ExoⅠ ranged from 0.1 Unit / μL (U / μL) to 20U / L, depending onfigure 1 It can be seen that within this range, the common logarithm (X) of ExoI concentration is linearly correlated with the ICPMS intensity signal (Y) of 63Cu. The linear equation is Y=2.23E4X +1.07E5, the correlation coefficient R 2 =0.9814, the limit of detection (LOD) was 0.029U / μL.
[0045] This example fully proves that the present invention can realize highly sensitive and label-free analysis...
Embodiment 2
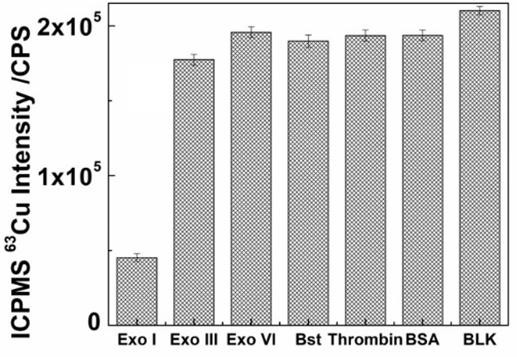
[0046] Example 2 Explore the specificity of the analytical method of the present invention to the detection of nuclease ExoI
[0047] In this example, taking Exo I as an example, several different common nucleases and proteins were detected by the label-free nuclease assay method based on PolyT-ssDNA template, and the specificity of the method was explored.
[0048] Using the above steps, use PolyT-ssDNA as a template to detect 5U / μL Exo I and 20U / μL Exo III, Exo VI, Bst, Thrombin, 0.05% m / v BSA respectively, keep the rest of the conditions exactly the same, and react at 37°C After 120 Min, 10 μL of 20 mM ascorbic acid solution and 35 μL of 1 mM copper sulfate solution were added and shaken at room temperature for 10 min. After magnetic separation, nitric acid digestion was performed, and stable isotope detection was performed by ICPMS. The result is as figure 2 As shown, except for Exo I, which can cause a significant decrease in signal, other common nucleases (exonuclease ...
Embodiment 3
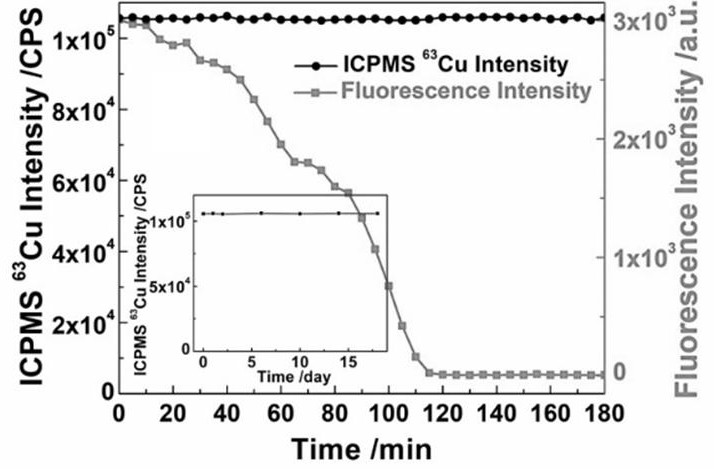
[0049] Example 3 Explore the long-term stability of the analytical method of the present invention for the detection of nuclease ExoI
[0050] In this example, Exo I was taken as an example to compare the signal stability under long-term repeated detection with the traditional label-free fluorescence detection method.
[0051] In the fluorescence detection experiment, PolyT-ssDNA was directly reacted with 1U / μL ExoⅠ, reacted at 37°C for 120Min, then added 10μL of 20mM ascorbic acid solution and 35μL of 1mM copper sulfate solution and shaken at room temperature for 10min to generate CuNPs with fluorescent properties. Fluorescence detection at an emission wavelength of 660 nm was performed at the excitation wavelength. The total detection time is 3 hours, and the interval between two times is 5 minutes. The intensity of each time is recorded and the relationship between fluorescence intensity and time is drawn.
[0052] In the experiment of stable isotope detection, PolyT-ssDNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com