CfDNA classification method and device and application
A classification method and a classification label technology, applied in the field of genomics and bioinformatics, can solve problems such as false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0175] Example 1: Preparation of cfDNA samples
[0176] 1. Target group
[0177] 95 cases of healthy people;
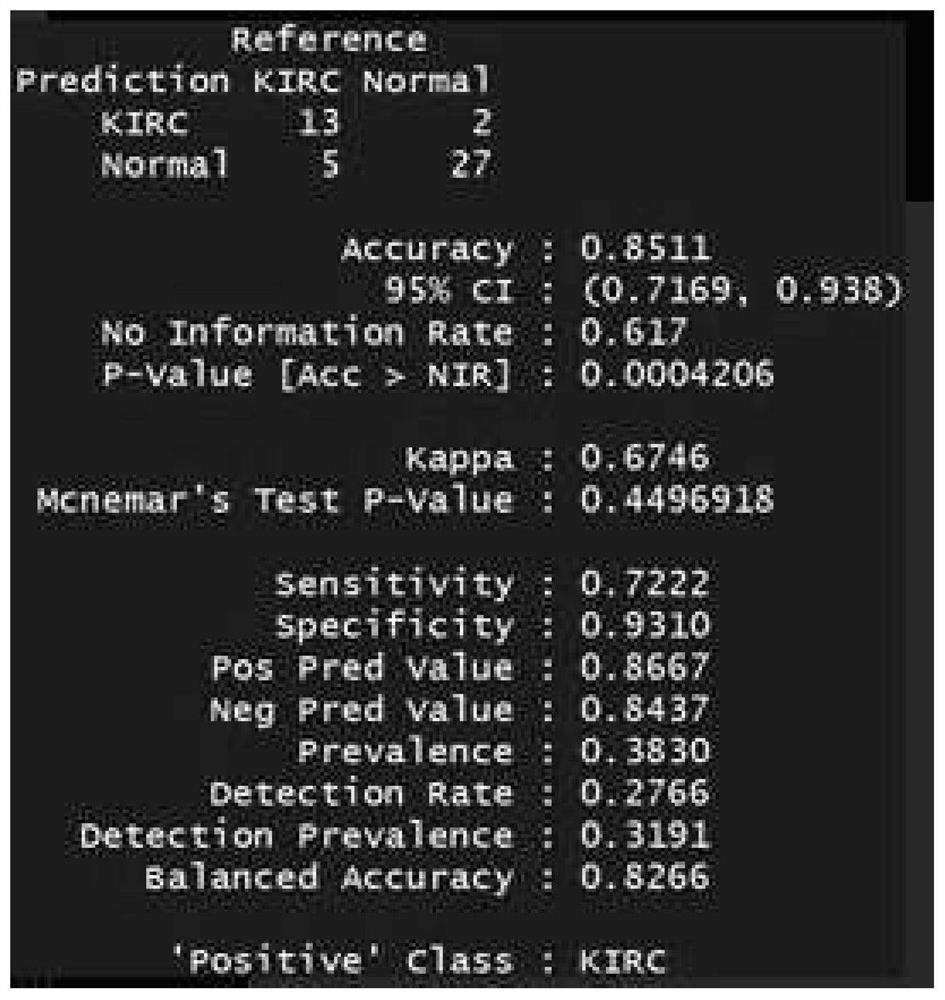
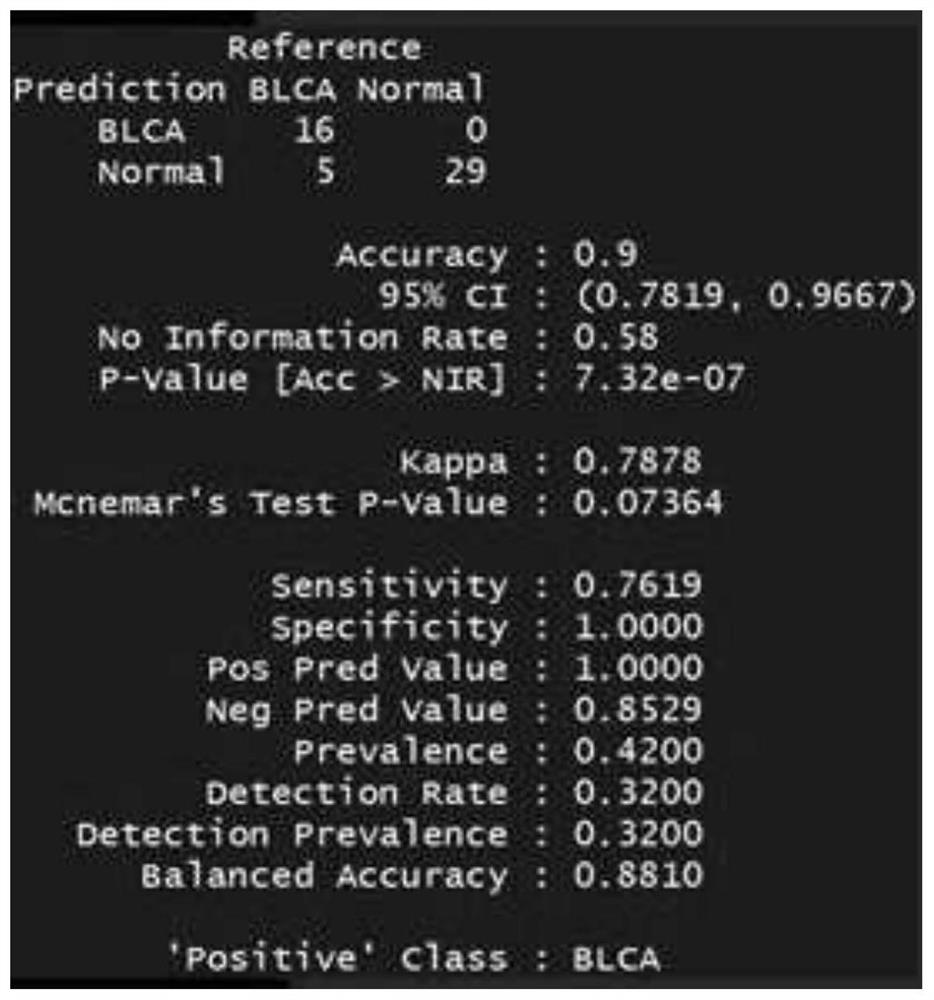
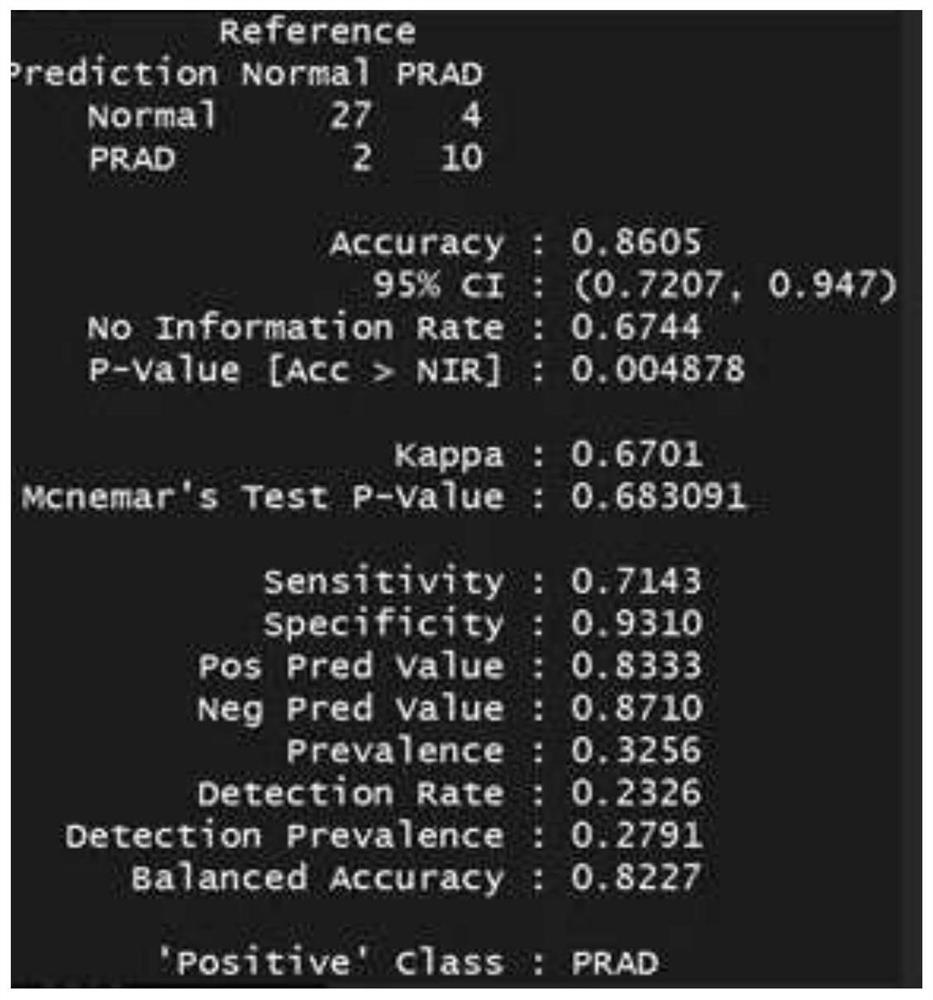
[0178] 172 patients, including: 58 patients with clear renal cell carcinoma (ccRCC), 69 patients with urothelial carcinoma and 45 patients with prostate cancer. Both were confirmed by biopsy of surgical samples.
[0179] A total of 267 cases of healthy people and patients.
[0180] 2. Experimental method
[0181](1) Collect the morning urine of the above-mentioned healthy people and the morning urine of tumor patients before operation. Each case of urine is collected in a 50ml centrifuge tube with a volume of about 20-50ml. Perform extraction to avoid cfDNA degradation.
[0182] (2) The collected morning urine samples were centrifuged at 3500 rpm for 15 minutes, and then the supernatants were taken respectively.
[0183] (3) Using zymo Quick-DNA TM The Urine Kit kit was used for the extraction of cfDNA. After extraction, the concentration was measured with ...
Embodiment 2
[0185] Example 2: Construction of whole genome library
[0186] 1. Experimental samples, reagents and instruments
[0187] 267 cases of cfDNA samples obtained in Example 1 above.
[0188] Urine cell-free DNA extraction kit: ZYMO Quick-DNA Urine Kit (ZYMO, Cat#: D3061).
[0189] Magnetic beads: AMPure XP beads (Beckman Coulter, Cat #: A63880).
[0190] Ordinary centrifuge.
[0191] 2. Experimental method
[0192] (1) Screen cfDNA of 100bp-300bp by magnetic beads (by controlling the ratio of the volume of magnetic beads to the volume of cfDNA samples, the range of the size of DNA fragments adsorbed by magnetic beads can be controlled). The specific operation is as follows:
[0193] Add 0.6 times the volume of magnetic beads to the extracted urine cfDNA, discard the magnetic beads after 5 minutes of adsorption, keep the supernatant, then add 0.3 times the volume of magnetic beads to the supernatant, discard the supernatant after 5 minutes of adsorption, and keep Magnetic ...
Embodiment 3
[0199] Example 3: HiSeq X10 system sequencing
[0200] 1. Reagents and Instruments
[0201] Samples to be tested: 267 libraries prepared in Example 2 above.
[0202] 2. Experimental method
[0203] Perform whole genome sequencing. Sequencing was commissioned by Novogene Sequencing Company.
[0204] 3. Experimental results
[0205] 150bp paired-end sequencing reads (pair-end reads) for each of the 267 libraries were obtained. The output sequencing depth of each sample is about 1X-5X. For subsequent tumor marker analysis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com