Dual-signal amplification probe, sensor, detection method and application
A dual signal amplification and detection sensor technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve problems such as aptamer sensors that are not combined to detect malathion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
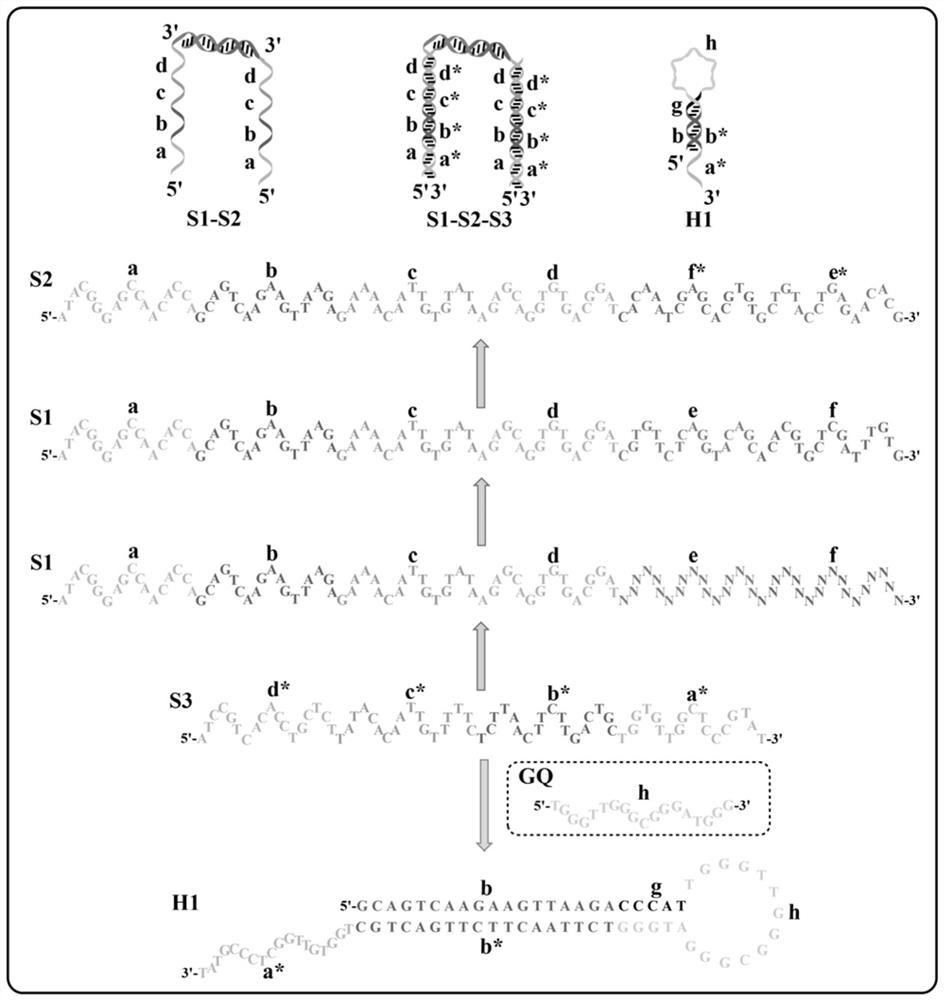
[0093]Example 1 Design of aptamer probe S1-S2-S3 and hairpin probe H1
[0094]In this embodiment, aptamer probes S1-S2-S3 and hairpin probe H1 are designed, including:
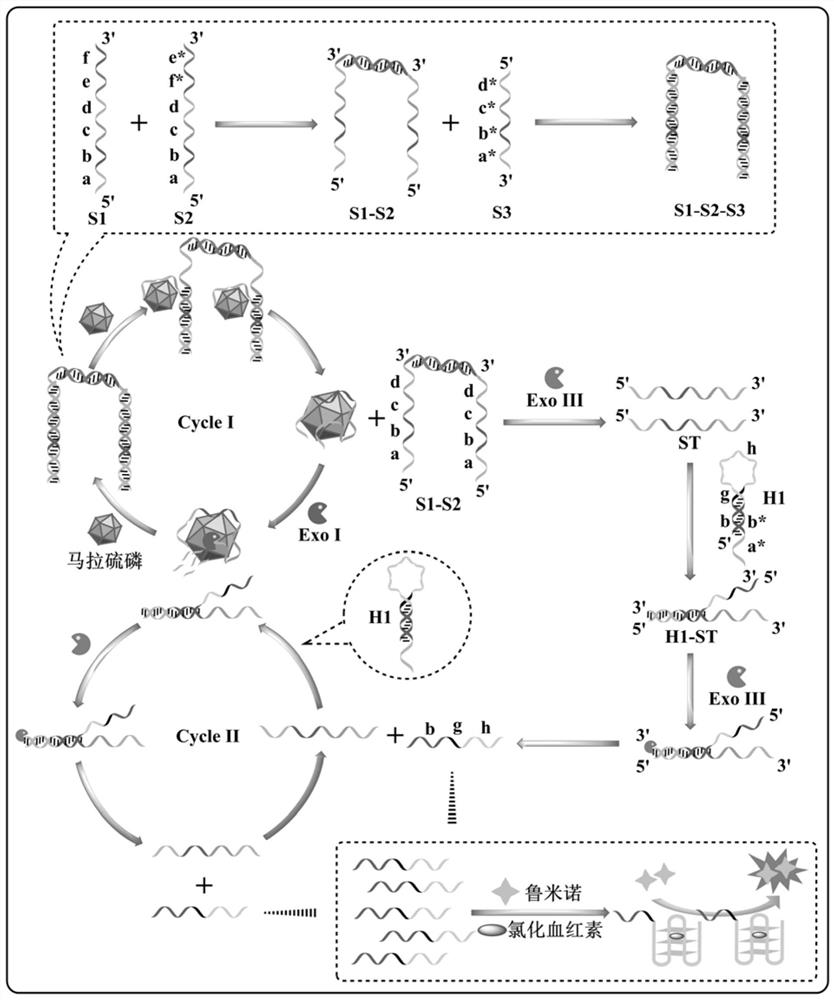
[0095]The aptamer probe is self-assembled by at least 3 single-stranded sequences, the single-stranded sequence includes sequence S1, sequence S2 and aptamer sequence S3 for binding to the target; said sequence S1 is from 5'to 3' The end includes a sequence ST complementary to the aptamer sequence S3 and a first binding sequence in sequence; the sequence S2 sequentially includes a sequence ST complementary to the aptamer sequence S3 and a complementary sequence of the first binding sequence from the 5'end to the 3'end;
[0096]The hairpin probe includes a second binding sequence, a G-quadruplex sequence, and a sequence ST* that can be partially complementary to the sequence ST from the 5'end to the 3'end; the second binding sequence and the G-quadruplex The 3'end sequence of the body sequence is partially complementary; the ...
Embodiment 2
[0112]This example provides a method for preparing the aptamer probes S1-S2-S3 and hairpin probes H1, H2, and H3 designed in Example 1.
[0113]The preparation method of hairpin probe H1: Centrifuge the oligonucleotide powder H1 at 12000 rpm (Ebend centrifuge 5417R) for 5 minutes, and then dissolve it in 20mM Tris-HCl buffer (200mM NaCl, 20mM KCl, 2mM MgCl)2, pH 7.4) prepared 10μM stock solution. The H1 stock solution was then heated to 95°C for 10 minutes, and then slowly cooled to room temperature to form a hairpin structure. Hairpin probes H2 and H3 were prepared in the same way.
[0114]Preparation method of aptamer probe S1-S2-S3: Centrifuge the oligonucleotide powders S1, S2 and S3 at 12000rpm for 5 minutes, and then dissolve them in 20mM Tris-HCl buffer (pH 7.4) to obtain 100μM stock solution solution. Then 10 μL of sequence S1 (100 μM), 10 μL of sequence S2 (100 μM) and 20 μL of sequence S3 (100 μM) were added to 60 μL of 20 mM Tris-HCl buffer (pH 7.4) and incubated at 37° C. for ...
Embodiment 3
[0115]Example 3 Double signal amplification detection kit
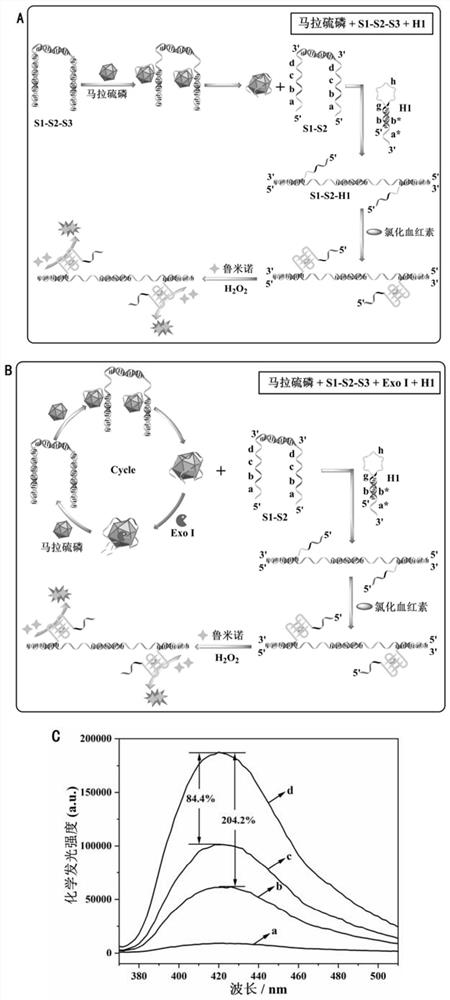
[0116]This embodiment provides a dual signal amplification detection kit, which includes the aptamer probe S1-S2-S3 and the hairpin probe H1 in the first embodiment.
[0117]Further, the molar ratio of the aptamer probe S1-S2-S3 to the hairpin probe H1 is 2:3.
[0118]Furthermore, the ratio of the amount of exonuclease Exo I, exonuclease Exo I and aptamer probe S1-S2-S3 is 15:0.02, and the ratio is U / nmol.
[0119]Further, it also includes exonuclease Exo III, the ratio of the amount of exonuclease Exo III and hairpin probe H1 is 20:0.03, and the ratio is U / nmol.
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com