Method for preparing porcine kidney epithelial cell line for stably expressing BE4 protein based on piggyBac transposon system
A porcine kidney epithelial cell, stable expression technology, applied in the field of genetic engineering and cell engineering, can solve the problems of affecting gene editing efficiency, low positive rate, low transfection efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1 piggyBac transposon and transposase expression vector construction
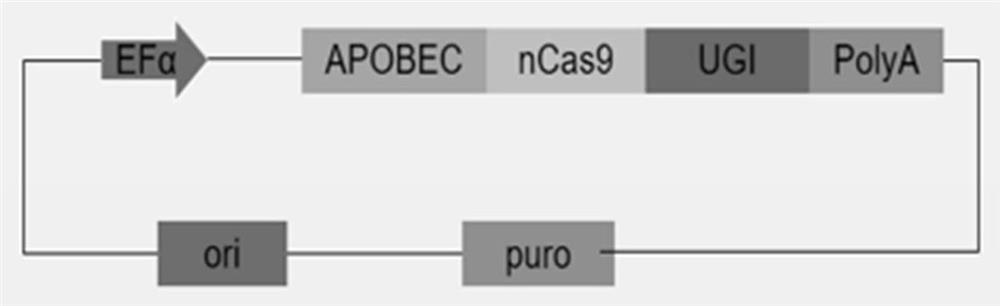
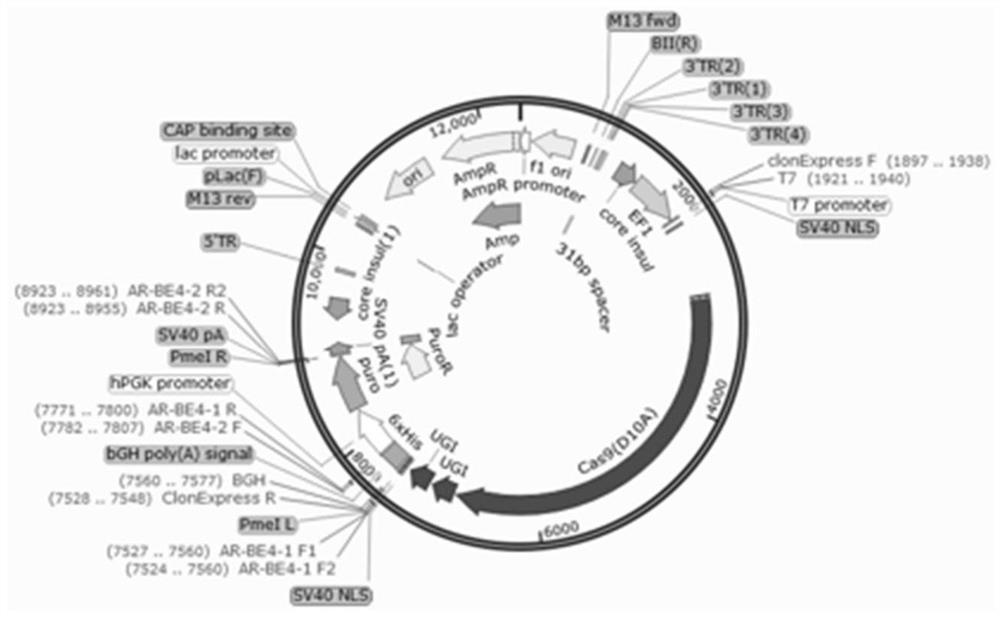
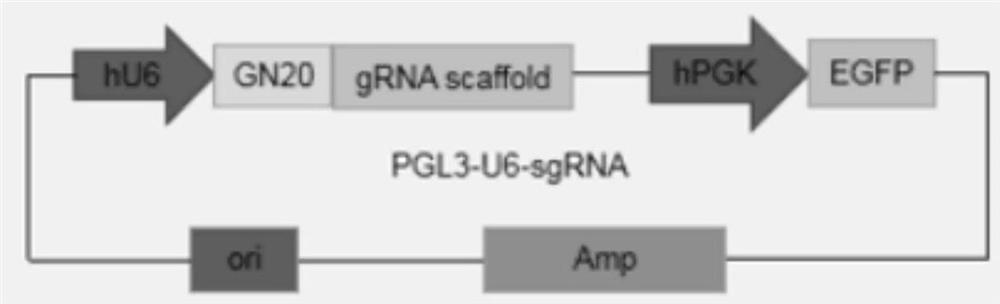
[0035] See Figure 1 with image 3 , piggyBac transposon carrier (PB-BE4max) includes elements such as BE4 fusion protein sequence, antibiotic selection marker gene (puromycin resistance gene), 5'TR and 3'TR homologous sequence, the sequence is as SEQ ID NO.1 As shown, the plasmid map as Figure 1B shown. The transposase carrier plasmid includes elements such as the transposase protein sequence, the sequence is shown in SEQ ID NO.2, and the plasmid map is shown in image 3 shown. The plasmid was positively transformed into Escherichia coli, expanded and cultivated, and then extracted by removing endotoxin to obtain the plasmid required for the experiment.
Embodiment 2
[0036] Example 2 PB-BE4max vector and transposase expression vector plasmid co-transfect PK-15 cells
[0037] The above-mentioned PB-BE4max vector and transposase expression vector plasmid were co-transfected into PK-15 cells, and the liposome transfection method (lipo2000) was used. The specific operation process was as follows:
[0038] Before transfection, the well-passaged PK-15 cells were inoculated into 6-well culture dishes, and transfected when the cells grew to a confluence of 70%-80%. According to the ratio of mass ratio PB-BE4max carrier: transposase expression vector = 2:1, the PB-BE4max vector and transposase expression vector were co-transfected into PK-15 cells, and operated according to the operation manual of lipo2000 (Invitrogen) . After the transfected cells were cultured for 48 hours, the medium was replaced with a fresh medium of 2 μg / mL puromycin, and the culture medium was continued at 37°C, 5% CO 2 Cultivate in a constant temperature cell incubator fo...
Embodiment 3
[0039]The picking of embodiment 3 monoclonal cell lines
[0040] refer to Figure 4 , dilute the cells of the positive cell population described in Example 2 to 7-8 cells / mL with fresh basal medium, then plate them in a 10cm dish, and cultivate them for 8-10 days; after the single cells grow into a single cell mass, Use the cloning ring to pick out the positive cell clusters and continue to expand the culture. Figure 4 Shown is a diagram of a monoclonal cell line formed from a single cell.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com