Stable polypeptide targeting SARS-CoV-2 spike protein and application of stable polypeptide
A technology of pneumonia virus and spike protein, applied in the field of bioengineering, can solve the problems of poor effect of new coronary pneumonia disease, and achieve the effect of broadening the scope of application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
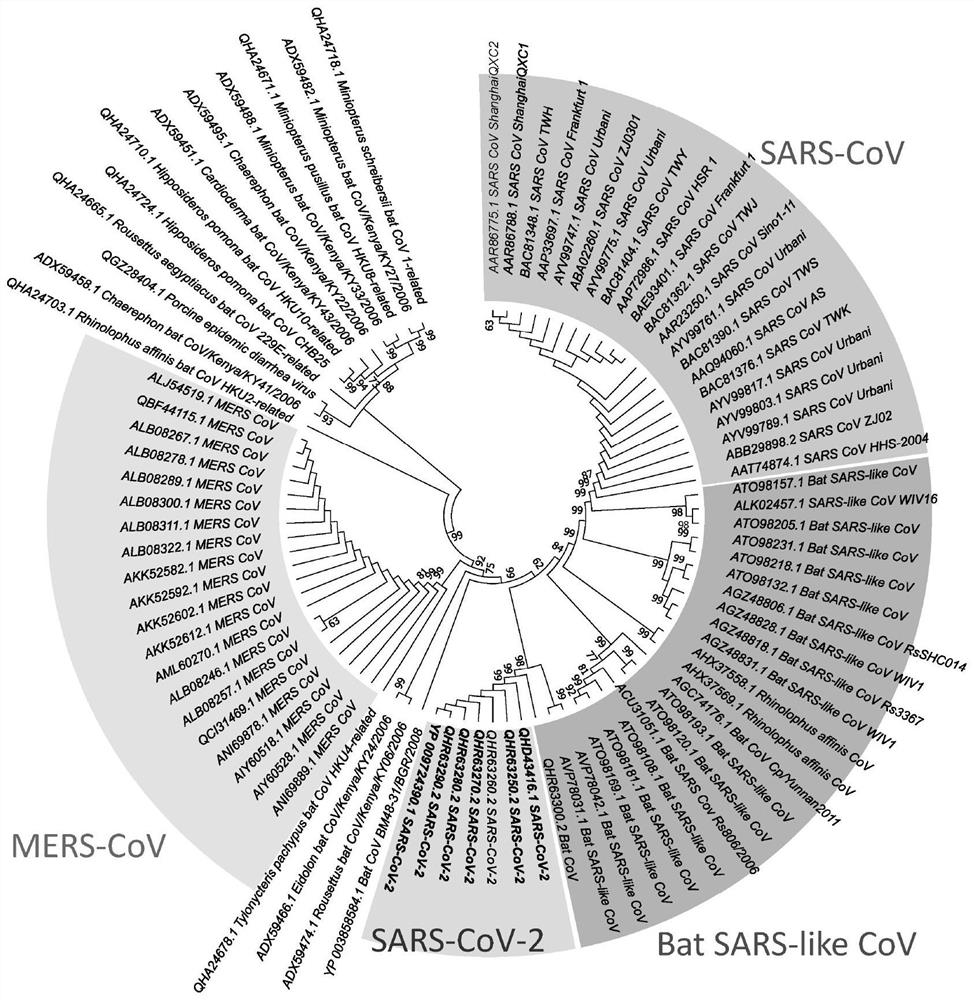
[0036] The present invention uses previous literature reports ((Zhao, Liu et al.2016, Tian, Yang et al.2017, Wang, Yu et al.2019)) to modify the polypeptide targeting the spike protein, through a section of known activity Sequence (Linker-IEEQAKTFLDKFNHEAEDLFYQS-CONH 2 ) structural transformation, it is expected to obtain an antiviral polypeptide targeting RBD with better druggability, higher stability and higher activity than the original polypeptide sequence.
[0037] In the modification process of the polypeptide, the selection of the polypeptide structure plays a key role, mainly the screening of its length. Scholars in the field have identified that in the 23 peptide, the first 7 amino acids play a decisive role and form a hydrogen bond network with ACE2. And the 8th-17th amino acid lacks the specific binding with human ACE2, and does not. In this study, we screened for the length of the linker. We synthesized three molecules with linkers of different lengths, as sho...
Embodiment 2
[0047] Preparation and separation and purification steps of the polypeptide of embodiment 2:
[0048] According to the amino acid sequence of solid-phase synthesis of polypeptides, the core steps of preparing the above stable polypeptides are as follows (taking TD-PROTAC as an example):
[0049]
[0050] The specific operation steps are:
[0051] (1) Polypeptide solid-phase synthesis: Weigh Rink amide MBHA resin into a peptide tube, add dichloromethane (DCM), and swell with nitrogen gas for 30 minutes. Add 50% (v / v) morpholine in N,N-dimethylformamide (DMF) solution, blow nitrogen gas for 30 minutes, and remove the Fmoc protecting group. After washing the resin alternately with DMF and DCM, the prepared Fmoc-AA-OH (5eq, 0.4M, DMF) solution, 6-chlorobenzotriazole-1,1,3,3-tetramethylurea Fluorophosphate ester (HCTU) (5eq, 0.38M, DMF) solution and N,N-diisopropylethylamine (DIPEA) (10eq) were mixed well, then added to the resin and blown with nitrogen for 1h. Take out the r...
Embodiment 3
[0056] Example 3 Expression of Spike Protein RBD Region
[0057] After RBD is expressed, it is deposited in the precipitate after broken cells. In this project, the RBD solution was obtained through the denaturation and renaturation experiments of RBD. Wherein, the buffer solution of denaturation is the guanidine hydrochloride of 6M of pH=7.2, and the buffer solution of refolding is the guanidine hydrochloride of 3M~0M of pH=7.2 ( Image 6 , Figure 7 ). However, the obtained protein has no activity, and the FP experiment shows that there is no obvious combination with the polypeptide.
[0058] In order to solve the problem that the prokaryotic expressed protein is inactive, we constructed an insect expression system for the RBD region of the spike protein and purified the corresponding protein. Figure 8 The gel image of the purified RBD protein is shown, and it can be seen that we obtained RBD protein with high purity through Ni column purification. We used the RBD prote...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com