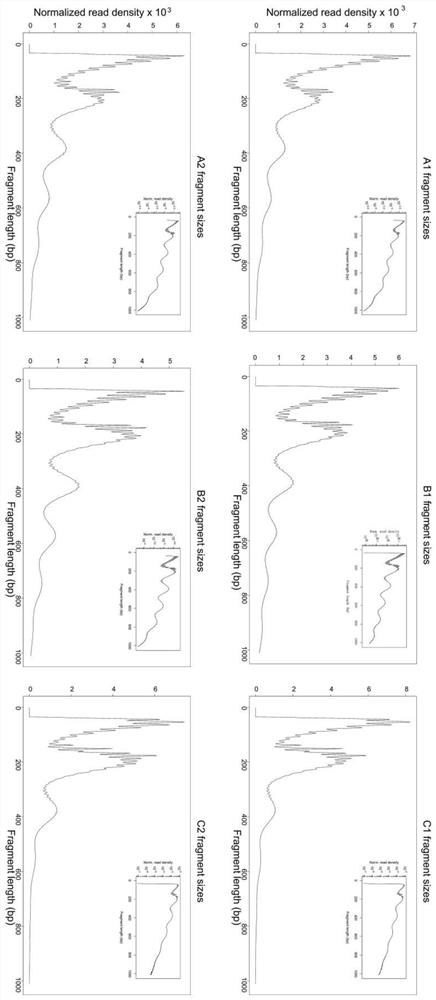
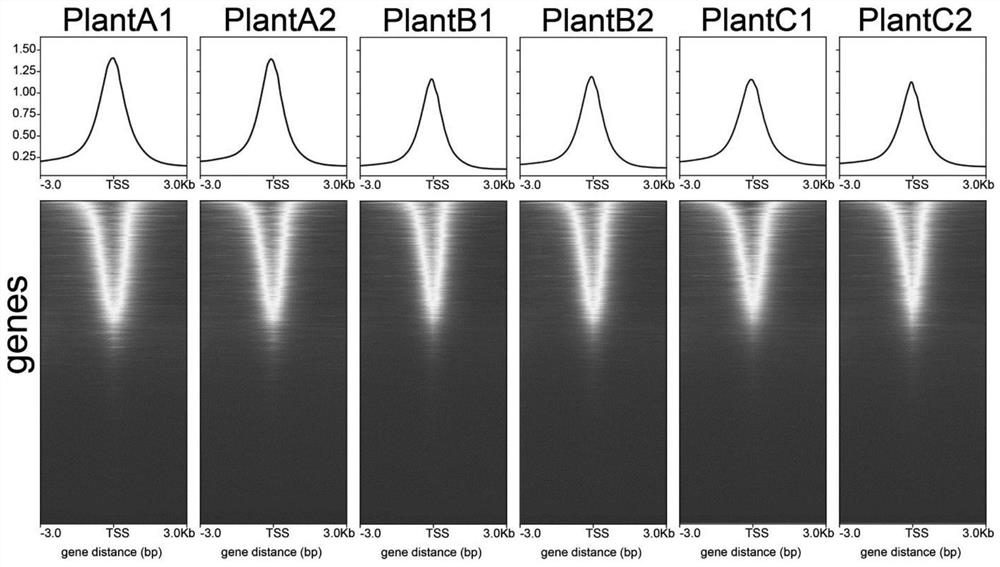
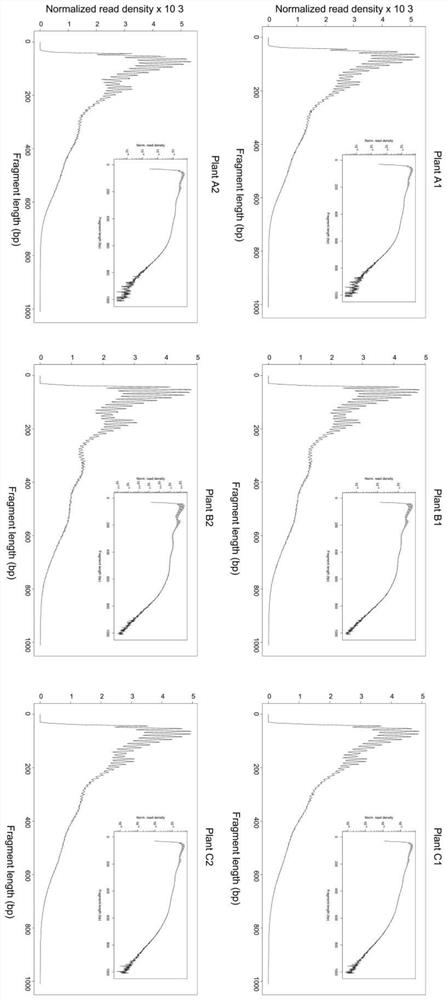
Preparation method of accurate quantitative ATAC-seq library and kit
A kit and library technology, applied in the field of accurate quantitative ATAC-seq library preparation methods and kits, can solve problems such as systematic errors and increased data repetition rate, and achieve the effect of accurate quantitative
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1: Plant samples
[0054] Preparation of the transposome complex:
[0055] (1) Thaw commercially purchased Tn5 Transposase on ice and shake well before use;
[0056] (2) The first adapter and the second adapter were annealed and hybridized with the single-stranded M to form a double-stranded adapter. The sequence of the single-stranded M was 5'-CTGTCTCTTATACACATCT-3, and the lengths of S2 and S6 were both 15 bp.
[0057] (3) Mix the Tn5 enzyme and the synthesized double-stranded adapter according to the molar ratio of 1:1, and incubate at room temperature for 1 hour to obtain the transposition complex.
[0058] Sequencing library preparation:
[0059] (1) Obtain Arabidopsis thaliana, wash it with ultrapure water for 3-5 times, wipe the surface of the leaf with a dust-free paper, use a pre-cooled mortar to crush the leaf tissue in liquid nitrogen, and freeze the leaf tissue Store in -80°C freezer for later use.
[0060] (2) Resuspend the tissue fragments ...
Embodiment 2
[0086] Example 2: Human samples
[0087] Preparation of the transposome complex:
[0088] (1) Thaw commercially purchased Tn5 Transposase on ice and shake well before use;
[0089] (2) The first adapter and the second adapter were annealed and hybridized with the single-stranded M to form a double-stranded adapter. The sequence of the single-stranded M was 5'-CTGTCTCTTATACACATCT-3, and the lengths of S2 and S6 were both 15 bp.
[0090] (3) The Tn5 enzyme and the synthesized double-stranded adapter were mixed according to the molar ratio of 1:1, and incubated at room temperature for 1 hour to obtain the transposition complex.
[0091] Sequencing library preparation:
[0092] (1) Obtain 10,000 intact human hepg2 cells, centrifuge at 500×g at 4°C for 5 minutes, and discard the supernatant.
[0093] (2) Add 50μl pre-cooled lysis buffer (10mM Tris-HCl, pH 7.4, 10mM NaCl, 3mMMgCl 2 , 0.1% IGEPPAL CA-630). Incubate on ice for 5-10min. Immediately centrifuge at 500×g 4°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com