Oligonucleotide aptamer specifically recognizing largemouth bass virus and its screening method and application
A technology of largemouth bass virus and oligonucleotide, which is applied in the field of oligonucleotide aptamer and its screening, can solve the problems of high cost, instability, long time required for antibody preparation, etc., and achieves low cost and easy operation. Simple and short screening period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
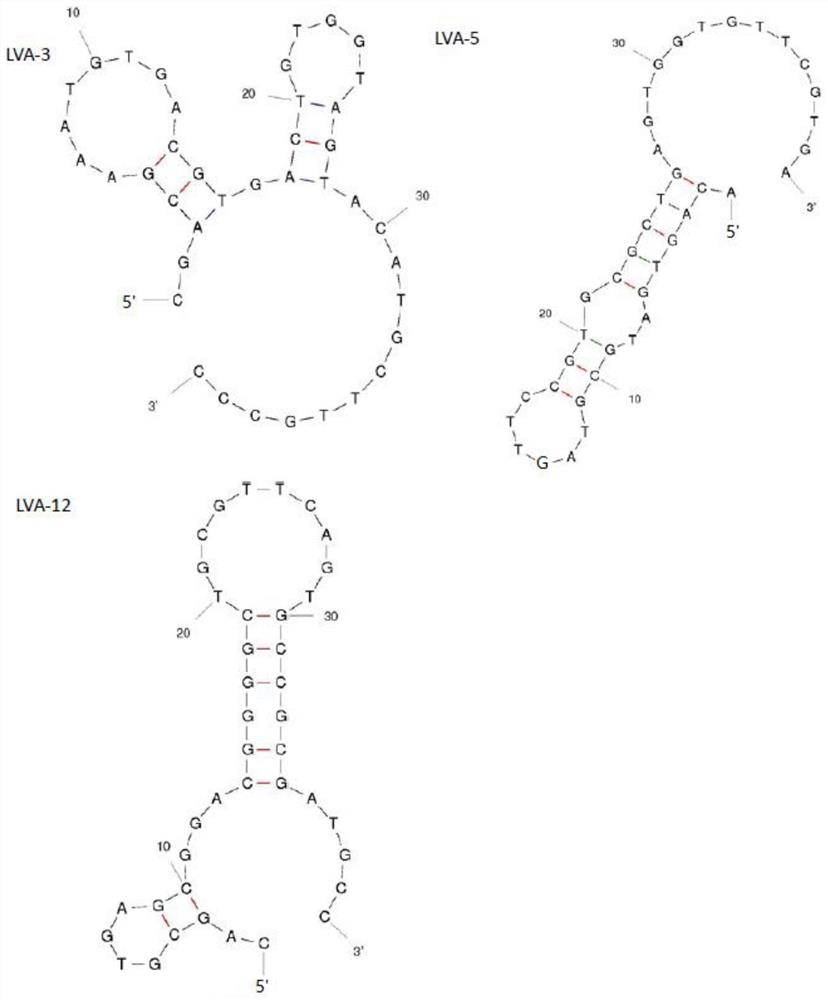
Image
Examples
Embodiment 1
[0044] The screening method of the nucleic acid aptamer that specifically recognizes largemouth bass virus comprises the following steps:
[0045] 1) Synthesis of random single-stranded DNA (ssDNA) library and primers
[0046] A random ssDNA library of single-stranded DNA with a length of 81nt was established: 5′-AGTATACGTATTACCTGCAGC(N 40)CGATATCTCGGAGATCTTGC-3′, with fixed sequences at both ends and a random sequence with 40 bases in the middle N40.
[0047] Upstream primer F: 5'-AGTATACGTATTACCTGCAGC-3';
[0048]Downstream primer R: 5′-GCAAGATCTCCGAGATATCG-3′; the random ssDNA library and primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd., and the random ssDNA library and primers were used ddH 2 O was prepared as a stock solution with a concentration of 10 μM and stored at -20°C for future use.
[0049] 2) Preparation of largemouth bass virus particles
[0050] After rinsing 50 mg of largemouth bass virus-positive liver with 0.0067M PBS buffer (pH7.4) f...
Embodiment 2
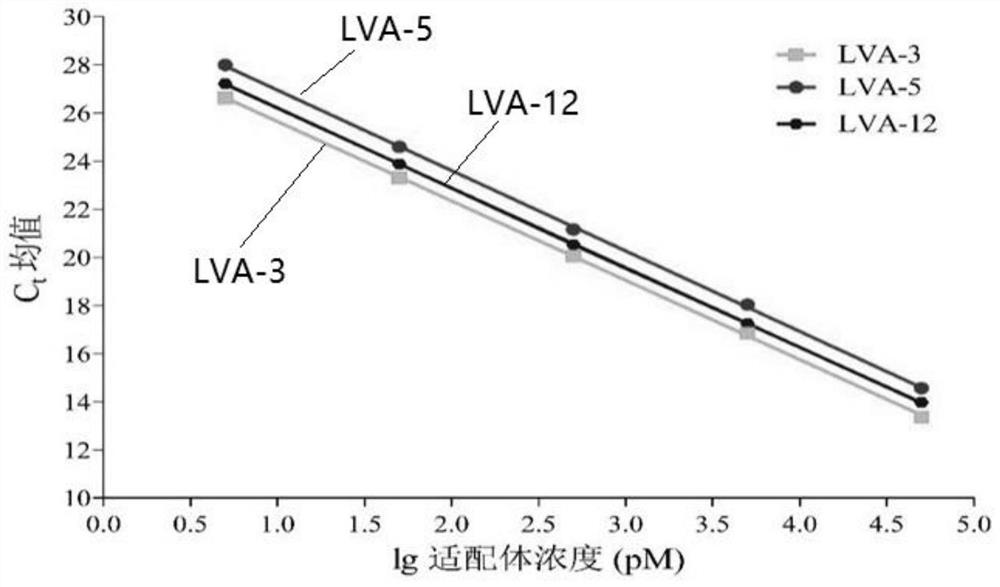
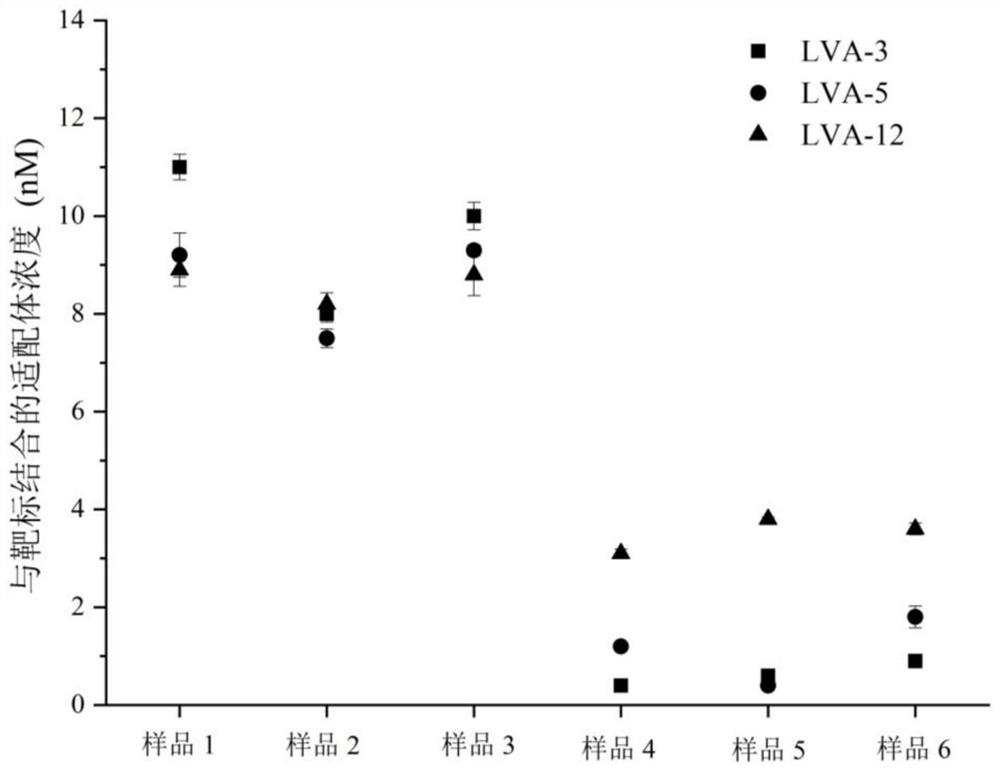
[0103] Eight diseased fish samples of largemouth bass were collected from different aquaculture farms in Zhejiang Province. The sample showed clinical signs of ulceration, exfoliation, superficial bleeding, and abdominal wall bleeding. Take 20 mg of liver from 8 samples of largemouth bass diseased fish, freeze-thaw and grind repeatedly, resuspend in 500 μL PBS, centrifuge at 8000 rpm for 5 min, and take the supernatant. Mix 50 μL of 40 nM LVA-3 nucleic acid aptamer aqueous solution with 50 μL of the above 8 supernatant samples, and incubate on a shaker at 25 °C and 100 rpm for 120 min; then add 50 μL of 15 mg / mL GO to the mixture and continue to incubate in the dark for 30 min , after centrifugation, the supernatant was taken for fluorescent quantitative PCR (the steps of fluorescent quantitative PCR here are the same as step 7 of the above-mentioned embodiment 1), and the Ct value was measured, thereby obtaining the concentration of aptamer bound to the target. The experimen...
Embodiment 3
[0105] 6 largemouth bass purchased randomly from the market were detected according to the method described in Example 2. Test results such as Figure 6 Shown: Sample 2 is LMBV positive, other samples are LMBV negative.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com