Method for analyzing selenium form of selenium-rich proteoglycan based on HPLC-ICP-MS
A technology of HPLC-ICP-MS and seleno-enriched protein, which is applied in the field of analyzing selenium-enriched proteoglycan selenium, can solve the problems of difficult control of acid extraction conditions, destruction of selenoamino acids, low efficiency, etc., and achieves good method stability, High recovery rate and accurate quantitative effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
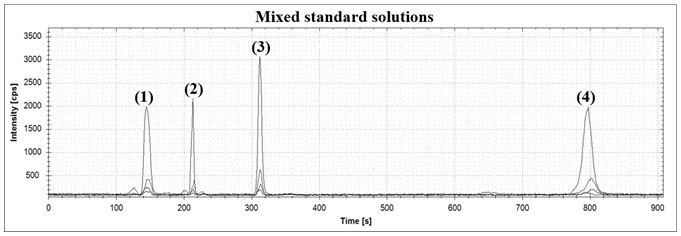
[0032] Embodiment 1 The drawing of standard curve
[0033] Accurately weigh 25.0 mg Na 2 SeO 3 Dissolve in water to make 50 µg / mL Na 2 SeO 3 Standard stock solution: Accurately weigh 20.0 mg of SeCys2, add it into water containing dilute acid to make a 200 µg / mL standard stock solution; accurately weigh 20.0 mg of SeMeCys, add it to water, and make a 200 µg / mL standard stock solution; accurately Weigh 20.0 mg of SeMet, add it into water, and make a standard stock solution of 200 µg / mL. Mix the above standard stocks in proportion so that Na 2 SeO 3 , SeCys2, SeMeCys and SeMet concentration ranges of 5-250 µg / L, 1-25 µg / L, 1-25 µg / L, 4-100 µg / L series of mixed standard working solutions. According to the conditions in Table 1, apply HPLC-IPC-MS analysis, and make a standard curve as shown in Table 2.
Embodiment 2
[0034] Example 2 Na in the sample 2 SeO 3 , SeCys2, SeMeCys and SeMet content determination
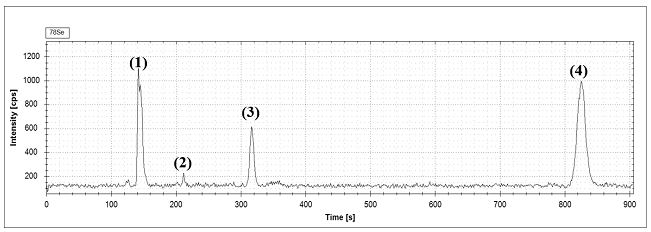
[0035]Accurately weigh 5 parts of about 15 mg of selenium-enriched proteoglycan and proteoglycan in a 2 mL centrifuge tube, add 5 mg of proteinase K, and then add 1.5 mL of Tris-HCl buffer (0.1 mol / L, pH=7.48), Shake at a constant temperature at 37°C for 18 hours (200 rpm), then centrifuge the enzymolysis solution for 10 min at 6000 rpm / min, collect the supernatant, and use HPLC-ICP-MS to analyze the selenium compound in the enzymolysis supernatant: Na 2 SeO 3 , SeCys2, SeMeCys and SeMet for analysis. See the hydrolysis diagram of selenium-enriched proteoglycan figure 2 , the content determination results are shown in Table 3.
Embodiment 3
[0036] Na in the sample of Example 3 2 SeO 3 、SeCys 2 , SeMeCys and SeMet recovery assay
[0037] Accurately weigh 3 parts of 15 mg Z0206 proteoglycan into a 2 mL centrifuge tube, add 4 mixed standard substances of low, medium and high according to Table 4, and repeat 5 times. Add 15 mg proteinase K and Tris-HCl buffer (0.1 mol / L, pH=7.48) to a final volume of 1.5 mL, shake at a constant temperature at 37°C for 18 hours (200 rpm), and then centrifuge the enzymatic solution 10min, the rotation speed is 6000 rpm / min, collect the supernatant, and apply HPLC-ICP-MS to the selenium compound in the enzymatic hydrolysis supernatant: Na 2 SeO 3 , SeCys2, SeMeCys and SeMet were analyzed and Na were calculated according to the standard curve 2 SeO 3 , SeCys2, SeMeCys and SeMet content and recovery.
[0038] According to the result and discussion that above embodiment 1-3 obtains:
[0039] Drawing of standard curve
[0040] to Na 2 SeO 3 , SeCys2, SeMeCys and SeMet to make a s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com