Multi-epitope fusion protein for preventing pseudomonas aeruginosa infection and encoding gene, expression vector and application thereof
A Pseudomonas aeruginosa and fusion protein technology, applied in the field of genetic engineering, can solve problems such as failure of vaccine development, achieve good immunogenicity and prevent infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
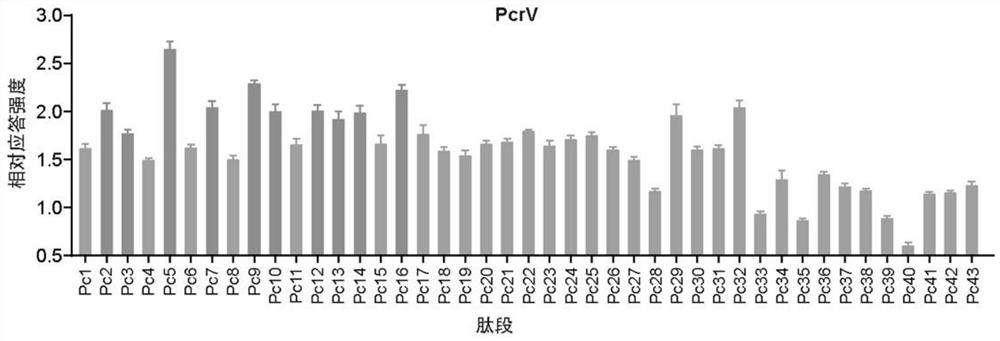
[0032] Example 1 Prediction analysis of antigenic protein dominant antigenic epitope
[0033] Neither the endpoints nor any values of the ranges disclosed herein are limited to such precise ranges or values, and these ranges or values are understood to include values approaching these ranges or values. For numerical ranges, between the endpoints of each range, between the endpoints of each range and individual point values, and between individual point values can be combined with each other to obtain one or more new numerical ranges, these values Ranges should be considered as specifically disclosed herein.
[0034] In a first aspect, the present invention provides a Pseudomonas aeruginosa vaccine recombinant protein, the vaccine recombinant protein is (a):
[0035] (a) a vaccine recombinant protein having the amino acid sequence shown in SEQ ID NO: 1;
[0036] According to the present invention, the recombinant protein of the Pseudomonas aeruginosa vaccine provided ...
Embodiment 2
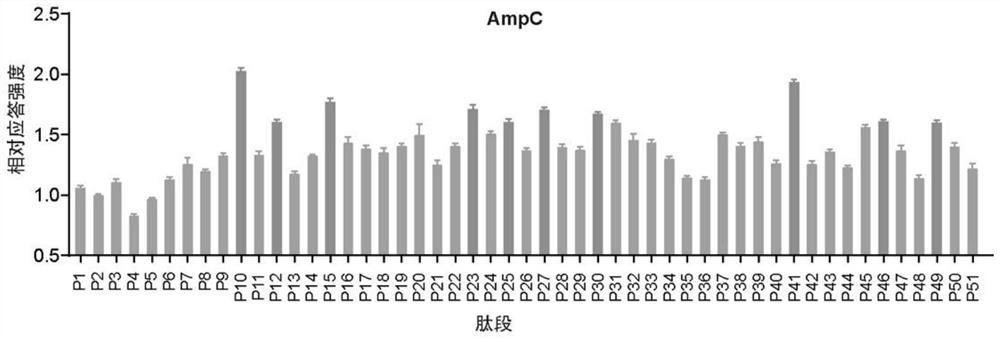
[0079] Example 2 Identification of epitopes that induce Th17 responses in AmpC
[0080] 1. Synthesis of multiple candidate epitope peptides
[0081] Jiangsu GenScript Biotechnology Co., Ltd. was entrusted to synthesize the walking polypeptide according to the amino acid sequence of AmpC (NCBI Reference Sequence: NP_252799.1). The following is the corresponding amino acid sequence of the synthetic peptide, see Table 3:
[0082] Table 3: Amino acid composition of AmpC peptide library
[0083]
[0084]
[0085]
[0086] 2. AmpC-immunized mice and isolation of lung lymphocytes after immunization
[0087]AmpC protein solution with a concentration of 5 mg / ml and Curdlan stock solution with a concentration of 20 mg / ml were mixed in equal volumes for preparation. After the mice were anesthetized with isoflurane gas, the above suspension was inoculated intranasally, 10ul per mouse, and the inoculation time points were 0, 14, and 21 days respectively. Fourteen days after th...
Embodiment 3
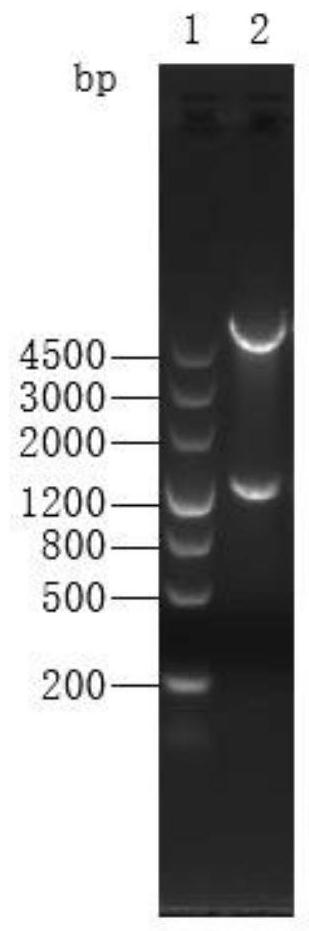
[0090] Example 3 Construction, expression, purification and identification of PVAC recombinant expression vector
[0091] 1. Construction of the carrier
[0092] Commissioned Shanghai Sangon Bioengineering Co., Ltd. to synthesize DNA encoding PVAC (SEQ ID NO:2)
[0093] 2. Transformation of recombinant plasmids
[0094] Take 1 tube of Escherichia coli XL-1blue competent cells from the -80°C refrigerator, and add 1 μl of the synthetic pGEX-6P-PVAC plasmid. Ice bath for 50min, heat shock in 42℃ metal bath for 60s, rapid ice bath for 1min. Add 1000 μl LB blank medium, mix well, and place in a shaker at 37° C. for 1 hour at 220 rpm. Each tube was centrifuged at 5000 rpm for 3 min at room temperature, 900 μl of the supernatant was discarded, and the bacteria were resuspended, and 100 μl was spread on an Amp-resistant LB plate. Pick well-separated colonies on the transformation plate, inoculate them in Amp-resistant LB medium, and culture them overnight at 37°C with shaking.
...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com