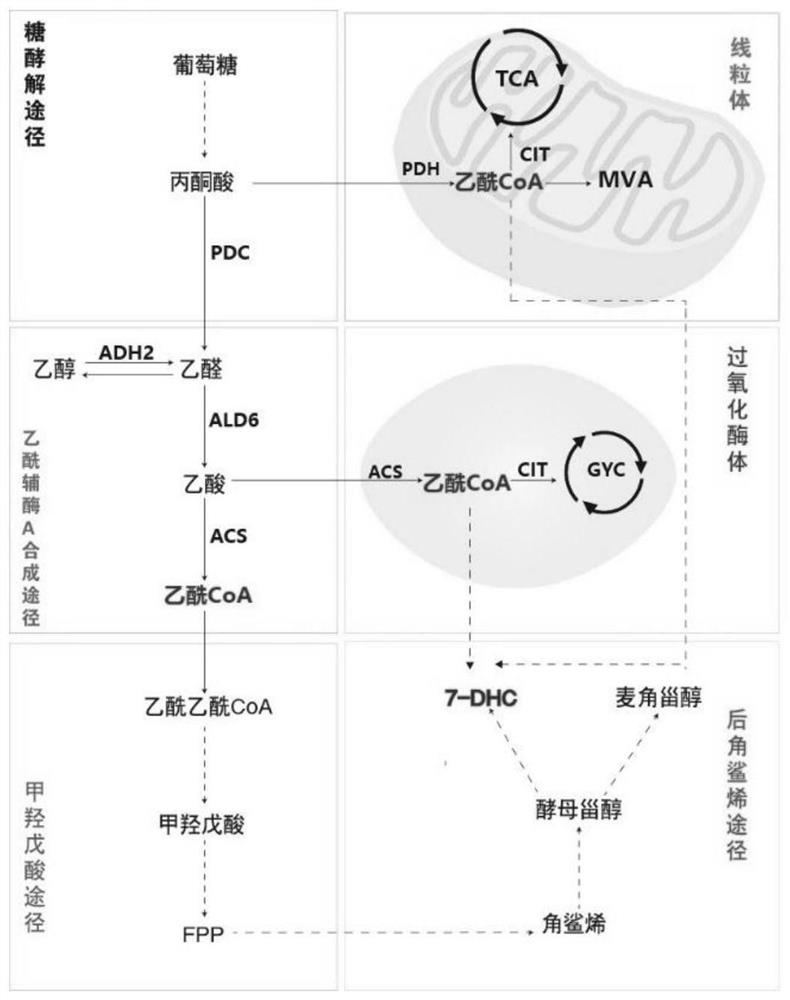
Method for increasing yield of 7-dehydrocholesterol in saccharomycetes by using compartmentalization
A dehydrocholesterol and yeast technology, applied in the field of metabolic engineering, can solve the problems of acetyl-CoA deficiency, reduce the feedback inhibition of metabolic pathways, increase the storage space of the final product, etc., achieve reduced losses, reduce feedback effects, and improve conversion efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
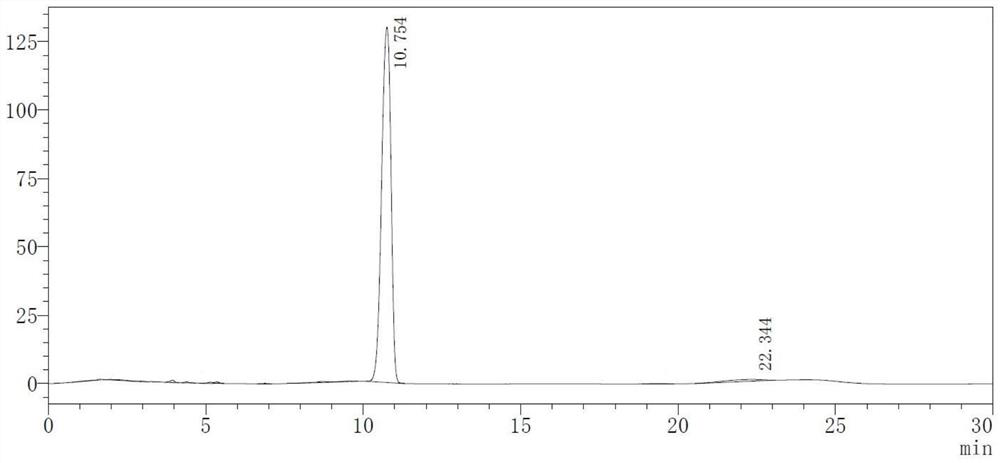
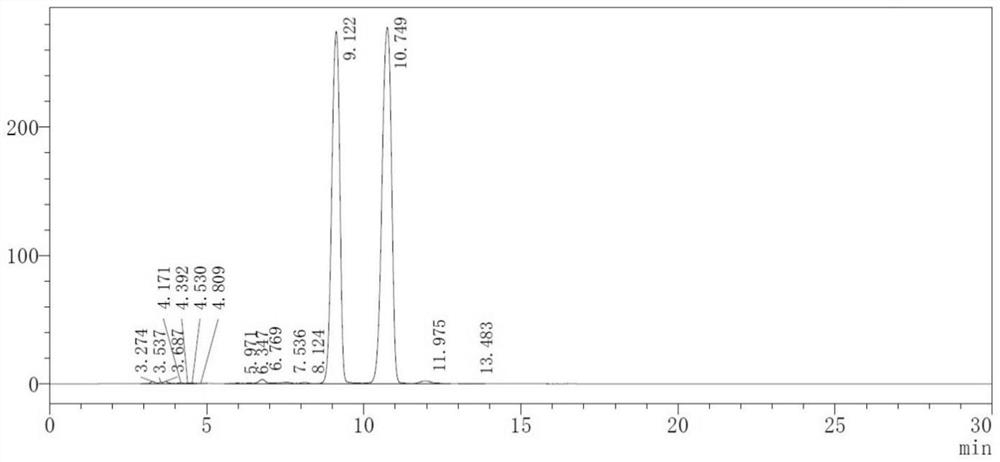
Image
Examples
Embodiment 1
[0025] Example 1: Construction of Saccharomyces cerevisiae CDHC-2
[0026] (a) Artificially synthesized gene fragment P TEF1 -DHCR24-T cyc1 -P GAP -DIC-T ADH1 , using the Saccharomyces cerevisiae S228C genome as a template, using primers 208F-F, 208F-R to amplify the gene fragment 208-F, using primers 208R-F, 208R-R to amplify the gene fragment 208-R, using pMHyLp-trp plasmid As a template, use primers loxT-F and loxT-R to amplify the loxT fragment of the gene fragment.
[0027] (b) the four fragments P obtained in step (a) TEF1 -DHCR24-T cyc1 -P GAP -DIC-T ADH1, 208F-F, 208-R and locxT gene fragments were subjected to overlap extension PCR, and after 1% agarose gel electrophoresis was verified to be correct, the fragments were recovered by cutting the gel to obtain the fusion gene fragment 208-F-loxT-P TEF1 -DHCR24-T cyc1 -P GAP -DIC-T ADH1 -208-R gene fragment.
[0028] (c) Transform the gene fragment obtained in step (b) into the competent strain of Saccharomyce...
Embodiment 2
[0039] Example 2: Construction of Saccharomyces cerevisiae mitochondrial compartmentalization
[0040] (a) Using the S228C genome of Saccharomyces cerevisiae as a template, the mitochondrial localization signal peptide MMF1 was selected. The gene fragment ERG12-MMF1 was amplified by primers ERG12-F and ERG12-MMF1-R, the gene fragment ERG13-MMF1 was amplified by primers ERG13-F1 and ERG13-MMF1-R1, and the gene fragment ERG13-MMF1 was amplified by primers GAL-F1 and GAL-R1. The GAL1 promoter and GAL10 promoter gene fragment GAL1-10 were amplified, the gene fragment Lox-Trp was amplified by primers loxT-GAL-F and LoxT-GAL-R, and the gene fragment was amplified by primers GAL80F1-F and GAL80F1-R Fragment GAL80F1, use primers GAL80R1-F, GAL80R1-R to amplify the gene fragment to obtain GAL80R1, use CYC1-S-F, CYC1-S-R to amplify the gene fragment to obtain CYC1, use primers ADH1-S-F, ADH1-S-R to amplify the gene fragment to obtain ADH1, Using MMF1-F, MMF1-R, MMF2-F, MMF2-R as primer...
Embodiment 3
[0067] Example 3: Construction of Saccharomyces cerevisiae peroxisome compartmentalization
[0068] (a) Using the S228C genome of Saccharomyces cerevisiae as a template, the peroxisome localization signal peptide PTS1 was selected. The gene fragment ERG12-PTS1 was amplified by primers ERG12-F and ERG12-PTS1-R, the gene fragment ERG13-PTS1 was amplified by primers ERG13-F and ERG13-PTS1-R, and the gene fragment ERG13-PTS1 was amplified by primers GAL-F and GAL-R. The GAL1 promoter and GAL10 promoter gene fragment GAL1-10 were amplified, the gene fragment Lox-Trp was amplified by primers loxT-GAL-F and LoxT-GAL-R, and the gene fragment was amplified by primers GAL80F3-F and GAL80F3-R Fragment GAL80F3, use primers GAL80R3-F, GAL80R3-R to amplify the gene fragment to obtain GAL80R3, use primers CYC1-F, CYC1-13-R to amplify the gene fragment to obtain CYC1, use primers ADH1-10-F, ADH1-R to amplify The gene fragment was obtained as ADH1.
[0069] (b) The CDHC-10 strain obtained in...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com