Single-cell single-tube sample preparation and single-cell proteomics analysis method
A proteomics and sample preparation technology, applied in the field of single-cell single-tube sample processing and single-cell proteomics analysis, which can solve problems such as low enzymatic hydrolysis efficiency, reduced enzymatic hydrolysis efficiency, and protein loss.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
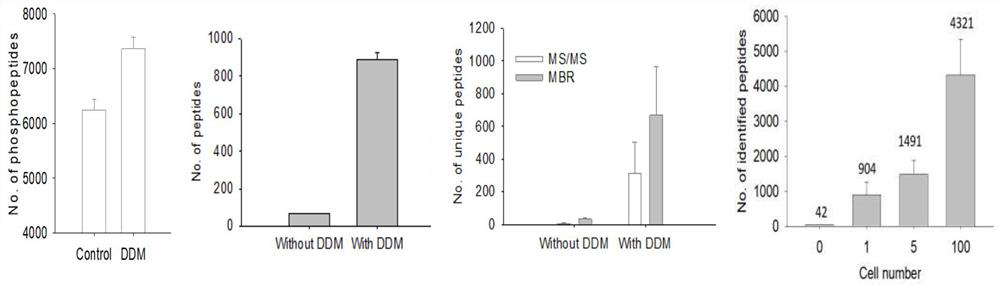
[0053] Example 1: DDM pretreatment research
[0054] (1): Multicellular research
[0055] To achieve precise quantification of the proteome of individual cells, we systematically assessed sample recovery using a homogeneous protein sample (i.e., cell lysate) in a single PCR tube.
[0056] Cell lysis: MCF cells in the growth phase were washed twice with ice-cold PBS solution, and 1ml of ice-cold PBS solution (containing 1% phosphatase inhibitor cocktail (Pierce), 10mM NaF (Sigma)) was added to collect the cells in a PCR tube. The cells were centrifuged at 1500rpm at 4°C for 10min, and the excess PBS solution was discarded. Then add cell lysis buffer (250mM HEPES, 8M urea, 150Mm Nacl, 1% Triton X-100, pH 6.0) to resuspend the cell pellet, the ratio of lysis buffer to cell pellet is 3:1. Centrifuge at 14000rpm at 4°C for 10min. Reserve Soluble protein component. Protein concentration was determined by BCA method.
[0057] Add 1 μg of cell lysate (approximately equal to 1000MC...
Embodiment 2
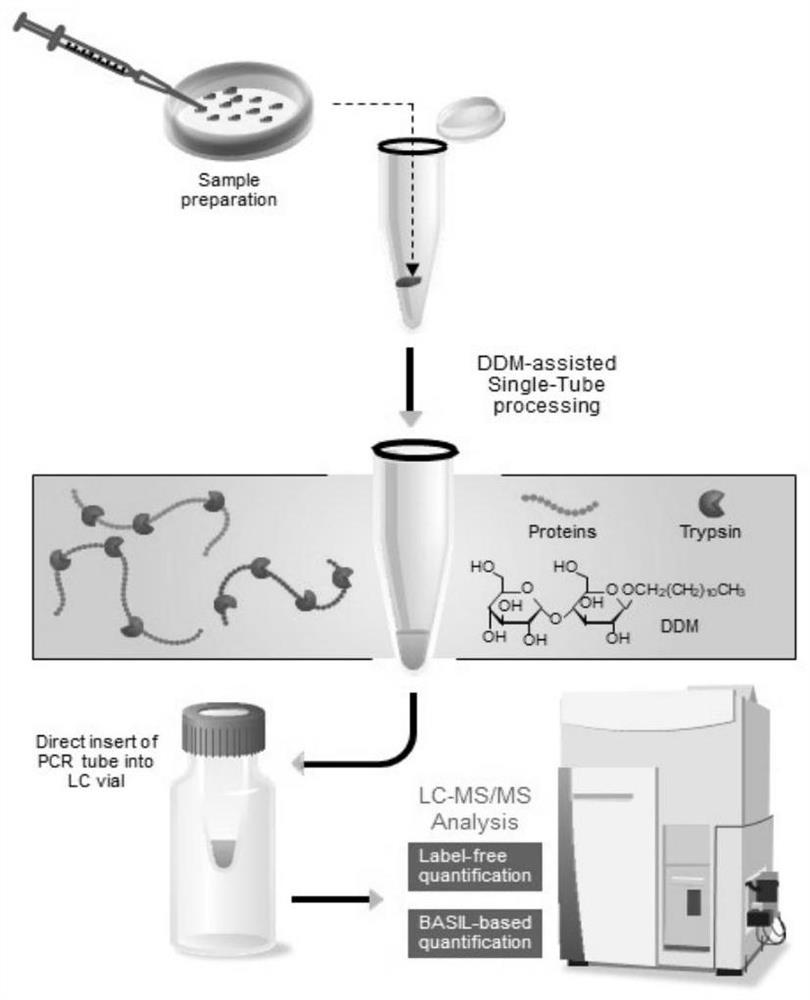
[0063] according to figure 2 Schematic diagram of the single-cell proteomics study on the human lung adenocarcinoma cell line Hcc827:
[0064] 1. Cell culture
[0065] Take out the cryopreservation tube containing the human lung adenocarcinoma cell line Hcc827, put it in a 37°C water bath, and shake it constantly to make it melt quickly; after the cells are thawed, take it out of the water bath, and centrifuge at 1000rpm for 5min; Remove the supernatant, add 1ml of fresh medium (1640+10% fetal bovine serum+1% sodium glutamate+1% sodium pyruvate) to resuspend; transfer to a culture bottle for static culture; cell culture at 37°C, 5% CO2 constant temperature and humidity incubator.
[0066] 2. Cell picking
[0067] Add 0.1% DDM to the PCR tube, fill it up completely (completely immerse the inner wall of the PCR tube), let stand at room temperature overnight (at least 12h), suck out the DDM solution, and air-dry the PCR tube naturally in a fume hood.
[0068] Select cancer c...
Embodiment 3
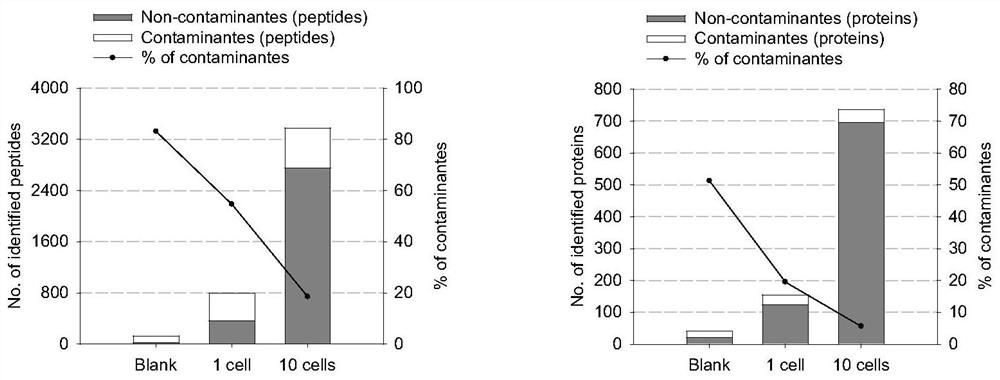
[0085]Compared with Example 2, the only difference is that during the cell picking process, 1, 5, 10, and 100H1975 cell samples were picked respectively for proteomics research (that is, the difference is only that the number of cells picked Different, other steps are with embodiment 2). For example, during the cell picking process, 1, 5, 10, and 100H1975 cells were picked from the fresh cell suspension using the serial dilution method and put into a single DDM-pretreated PCR tube. The sample in the sample was digested by enzyme, and the PCR tube was capped and inserted into a 2ml sample bottle for LC-MS / MS analysis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com