Single nucleotide mutation site SNP and KASP markers significantly associated with soybean protein content and application thereof
A soybean protein and mutation site technology, applied in the field of molecular genetics and breeding, can solve the problems of time-consuming, labor-intensive, low, susceptible to environmental interference, accuracy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
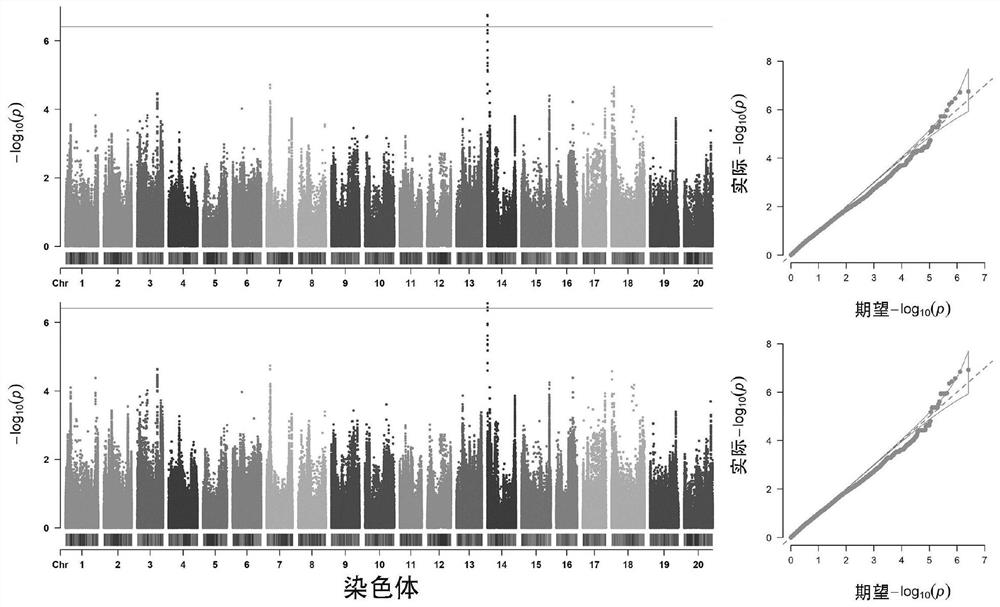
[0040] Example 1: Acquisition of Nucleotide Mutation Sites (SNPs) Significantly Associated with Soybean Protein Content
[0041] (1) Determination of protein content: Take the dried laboratory soybean samples and mix them thoroughly, then crush them, and pass all the grains through a 0.25mm aperture sieve, and put them into sample bottles for later use. Weigh 0.2g of the sample, accurate to 0.0001g, and place it in a 300mL digestion tube. Then the protein was determined according to the method of national standard GB 5009.5-2016.
[0042] (2) DNA extraction and high-throughput sequencing: The CTAB method was used to extract the genomic DNA of 264 young soybean leaves, and the whole genome was resequenced.
[0043] (3) Genome-wide association analysis (GWAS): Using the GAPIT algorithm package in the R language software, the calculation model is a mixed linear model (MLM), GWAS analysis detected 24 SNP sites significantly associated with soybean protein content, and all Locate...
Embodiment 2
[0044] Example 2: Development of KASP marker-specific primers
[0045] Using the Primer-BLAST function of NCBI (https: / / www.ncbi.nlm.nih.gov / ), three primers were designed according to the sequence of Seq ID NO.1, upstream primer F1 (Seq ID NO.2), upstream primer F2 ( Seq ID NO.3) and downstream primer R (Seq ID NO.4), wherein F1 and F2 respectively contain FAM and HEX fluorescent linker sequences (underlined), the sequences are as follows:
[0046] F1 sequence: 5'- gaaggtgaccaagttcatgct gttgaagtgagatatggtggacg-3'
[0047] F2 sequence: 5'- gaaggtcggagtcaacggatt gttgaagtgagatatggtggaca-3'
[0048] R sequence: 5'-ttccacctcgccattcatcc-3'
Embodiment 3
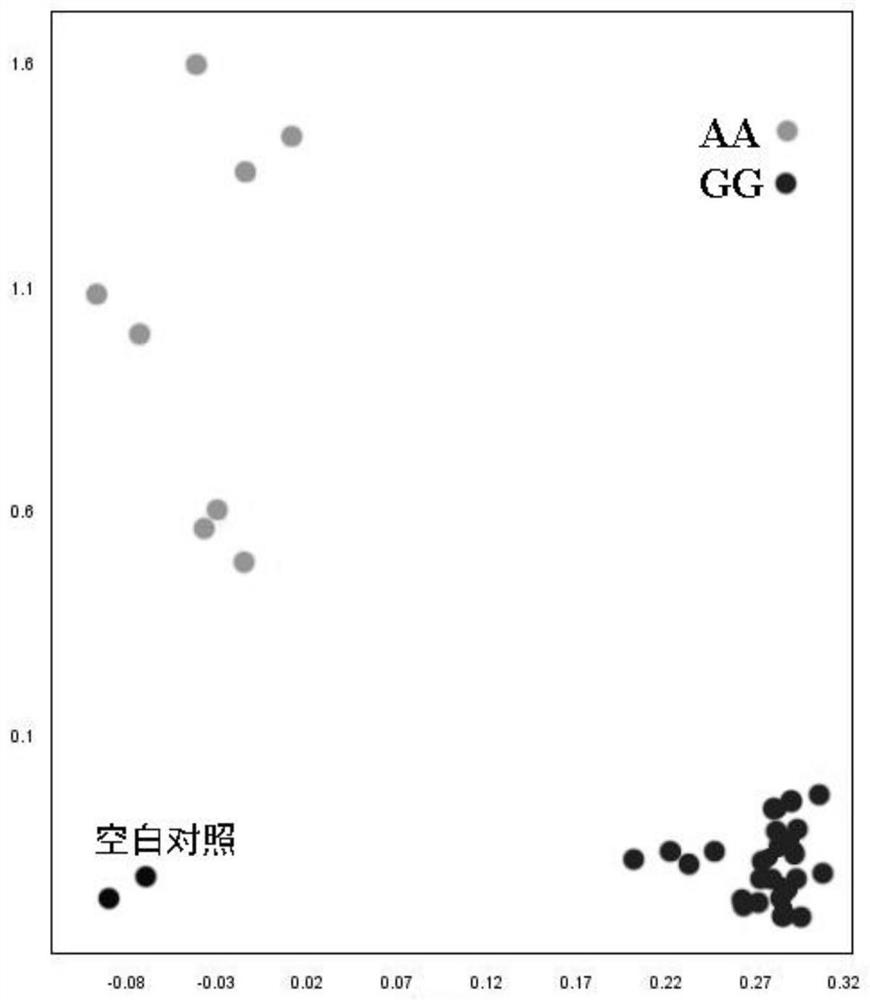
[0049] Example 3: Detecting genotypes of different varieties of soybean SNP sites and its application
[0050] The genomic DNA of the soybean samples is extracted respectively, and the genomic DNA is used as a template to carry out PCR amplification with KASP labeled special primers to obtain PCR amplification products. PCR amplification was carried out in the ABI7500 real-time fluorescent quantitative PCR instrument. After PCR, the instrument can perform genotyping according to the fluorescent signal. The amplification systems described are all 10 μl reaction systems: soybean sample DNA template, 25ng / μl, 2 μl; 2×KASP Master mix 5 μl; KASPassay Mix, F1:F2:R=2:2:5, 0.14 μl; water 2.9 μl. The reaction conditions included pre-denaturation at 94°C for 15 min; denaturation at 94°C for 20 sec, annealing at 61-55°C for 60 sec, 10 cycles with each cycle lowering 0.6°C; denaturation at 94°C for 20 sec, annealing at 55°C for 60 sec, 26 cycles.
[0051] After the reaction is completed...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com