Zearalenone degrading enzyme mutants with improved enzyme activity as well as coding gene and application of zearalenone degrading enzyme mutants
A technology of zearalenone and encoding gene, which is applied in the fields of genetic engineering and enzyme engineering, can solve the problems of narrow substrate application range, low catalytic activity, low enzyme production level, etc., achieves good application potential, improved degradation efficiency, The effect of good market application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Error-prone PCR construction and screening of zearalenone-degrading enzyme mutant library
[0050] refer to Clonostachys rosea The amino acid sequence (SEQ ID NO: 1) and its optimized DNA sequence (SEQ ID NO: 2) of the source zearalenone degrading enzyme were artificially synthesized, and the EcoR I restriction enzyme was designed at the 5' end Cutting site, Not I restriction enzyme cutting site was designed at the 3' end. The synthesized gene fragment was double digested with EcoR I and Not I, recovered by gel cutting, connected to the pET-21a(+) vector, transformed into Escherichia coli DH5α, positive clones were screened on the ampicillin-resistant LB plate, and the recombinant was verified by sequencing Vector pET-YMDS0, recombinant bacteria BL21-YMDS0.
[0051] Use the GeneMorph II random mutation PCR kit to use the gene shown in SEQ ID NO: 2 as a template for random mutation. The primer sequences used are as follows:
[0052] YMDS-F: A GAATTC ATGCG...
Embodiment 2
[0059] Example 2: Expression of Zearalenone Degrading Enzyme Mutant in Pichia pastoris GS115
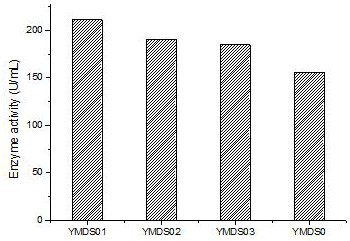
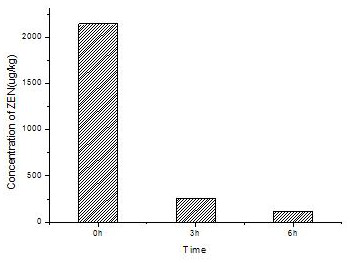
[0060] The artificially synthesized gene YMDS0 and the zearalenone degrading enzyme mutants YMDS01, YMDS02, and YMDS03 screened in Example 1 were constructed on the pPIC9K plasmid to obtain the expression vectors pPIC-YMDS0, pPIC-YMDS01, pPIC-YMDS02, pPIC-YMDS03; the expression vectors of each mutant were digested with Sal I and linearized, then electrotransformed into Pichia pastoris GS115, and the transformants of the expression strains of each mutant were screened on the MD plate and transformed into Received into YPD plate for activation.
[0061] The activated transformants were picked and inoculated in BMGY medium, cultured with shaking at 30°C for 18 hours, and then centrifuged to obtain bacterial cells. Transfer an appropriate amount of bacteria into BMMY medium to make the concentration of bacteria reach OD600=1, continue shaking culture, and add 1% methanol of the culture ...
Embodiment 3
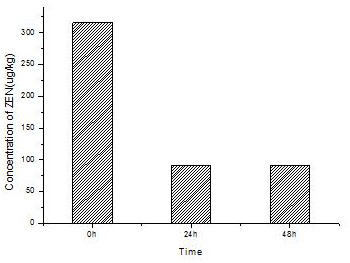
[0062] Embodiment 3: Fermentation and preparation of zearalenone degrading enzyme mutant in 30L fermenter
[0063] Streak the Pichia pastoris preserved in the glycerol tube on the YPD plate, and culture it upside down at 30°C for 3 days to grow a single colony. Pick a single colony that grows well and continue to streak on the YPD plate to activate the Pichia obtained from the third generation. A single colony of red yeast was inoculated in 50 mL of BMGY medium, and cultured at 30°C and 200 rpm for 24 hours. Inoculate into 1000mL BMGY medium with a 2% inoculum amount, cultivate at 30°C and 200rpm until the OD600 is 5, and use it as seed solution to inoculate the fermenter.
[0064] Fermentation production process: BSM medium, pH 4.8, temperature 30°C, stirring rate 500 rpm, ventilation rate 1.5 (v / v), dissolved oxygen controlled above 20%.
[0065] The fermentation process is divided into three stages: (1) Bacteria culture stage: 8% of the seed liquid is added, and cultured a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com