Preparation method for synthesizing microbial self-luminescence biosensor by using self-luminescence operon and corresponding biosensor and application
A biosensor and self-luminous technology, applied in the fields of genetic engineering and molecular biology, can solve problems such as complexity, interference signals, and unfriendly applications, and achieve the effects of simple operation, high sensitivity, and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1: the acquisition of gene and the construction of vector
[0040] 1. Acquisition of genes
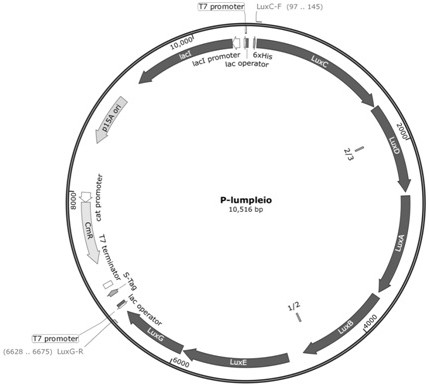
[0041]A luxABCDE operon derived from Photobacterium leiognathi (Photobacterium leiognathi), whose nucleotide sequence is shown in SEQ ID NO.1, was chemically synthesized onto the pUC-57 vector by BGI Corporation to obtain the pUC-lumPleio vector .
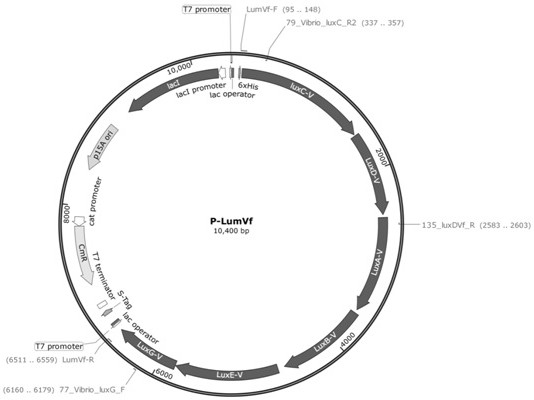
[0042] A luxABCDE operon derived from Aliivibrio fischeri, whose nucleotide sequence is shown in SEQ ID NO.2, was chemically synthesized by Huada Gene Company onto the pUC-57 vector to obtain the pUC-lumVf vector .
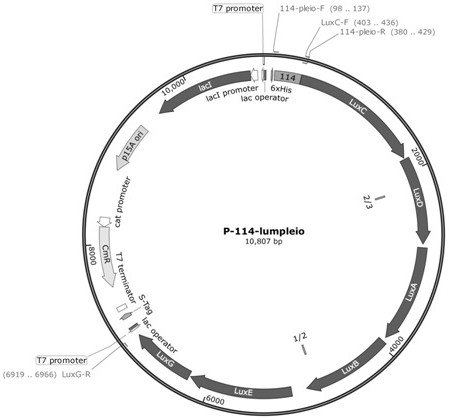
[0043] The 114 promoter gene, whose nucleotide sequence is shown in SEQ ID NO.3, was chemically synthesized by Huada Gene Company into pUC-57 vector to obtain pUC-114 vector.
[0044] 2. Construction of p-lumPleio and p-lumVf expression vectors
[0045] (1) Using pUC-lumPleio as a template, primer LuxC-F and primer LuxG-R, perform polymerase chain reaction (PCR) to amplify the lumPleio fragment. The PCR am...
Embodiment 2
[0105] Embodiment 2: the construction of biosensor
[0106] The p-114-lumPleio recombinant plasmid was transformed into Escherichia coli BW25113 competent cells (purchased from Weidi Biotechnology, product number DL2050), spread on the LB solid plate containing 34 mg / L chloramphenicol, and obtained positive clones by PCR screening, thus The engineering strain XLUM1 containing the vector p-114-lumPleio was obtained.
[0107] The p-114-lumVf recombinant plasmid was transformed into Escherichia coli BW25113 competent cells (purchased from Weidi Biotechnology, product number DL2050), spread on the LB solid plate containing 4 mg / L chloramphenicol, and obtained positive clones by PCR screening, thus The engineering strain XLUM2 containing vector p-114-lumVf was obtained.
Embodiment 3
[0108] Embodiment 3: The application of microbial self-luminescence biosensor to detect explosive molecules
[0109] 1. Strain activation and cultivation
[0110] The engineering strains XLUM1 and XLUM2 with correct sequencing were respectively transferred to LB liquid medium containing 34 mg / L chloramphenicol, and cultured overnight at 37°C to obtain bacterial liquid.
[0111] Add 330 μL 60% glucose, 10 μL 1 M magnesium sulfate stock solution, and 200 μL bacterial solution to 10 mL of M9 liquid medium containing 34 mg / L chloramphenicol, culture on a shaker at 37 ° C, and grow to OD 600 = about 0.2.
[0112] 2. Preparation of 2,4-DNT solution
[0113] Prepare 20mg / mL mother solution 2,4-DNT (100mg2,4-DNT dissolved in 5mL absolute ethanol);
[0114] Prepare the diluted 2,4-DNT solution in the following proportions:
[0115] 100mg / L: 5μL stock solution + 980μL M9 medium + 15μL absolute ethanol;
[0116] 50mg / L: 2.5μL mother solution + 980μL M9 medium + 17.5μL absolute ethan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com