Application of gene TSA in detection of collybia albuminosa derived components
A technology of derived components and genes, applied in the field of biological species identification and PCR detection, to achieve the effect of less time-consuming, good specificity and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Embodiment 1: Common internal standard gene of chicken fir source TSA screening
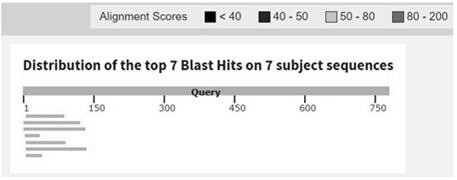
[0017] In the Nucleotide database of NCBI, search for the gene information of cyprinus fir, and finally screen out the genes used to encode glycosidase through BLAST comparison analysis. TSA Gene (accession number: JO124034.1) was used as an internal standard gene, TSA BLAST results and details for genes are in figure 1 , TSA The BLAST alignment results of the gene show that there is no complete sequence of any other gene with TSA The sequences of the genes are identical or similar, which means that TSA The gene has no homology with other genes and has strong species specificity.
Embodiment 2
[0018] Embodiment 2: Verification of the universal internal standard gene of the origin component of chicken fir
[0019] 1. For the internal gene screened in Example 1 TSA , use the Primer Premier 5.0 software to design primers, and perform BLAST comparative analysis on the designed primers to ensure the specificity of the primers. The primer sequences are shown in Table 1; the amplified sequence is the 213th base to the 1st base of SEQ ID NO:1 546 bases;
[0020] Table 1 TSA Gene PCR primer sequence
[0021] ;
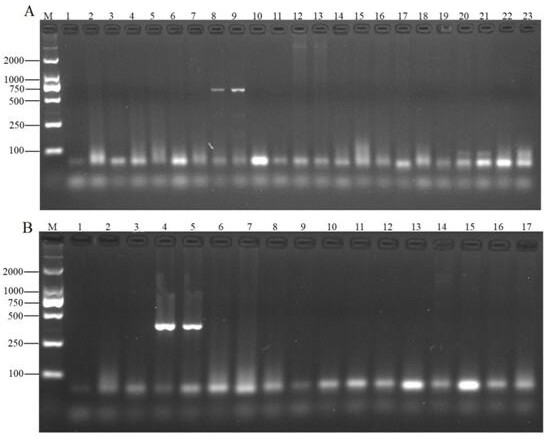
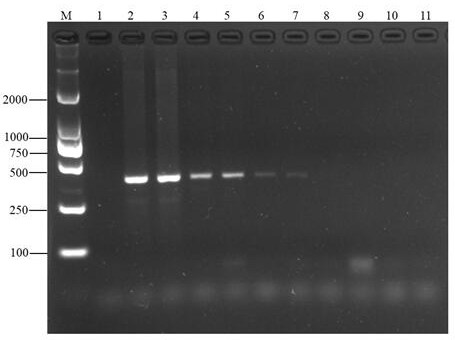
[0022] 2. Qualitative verification of the species-specificity of the standard gene primers in C. fir
[0023] Yellow boletus liver, northern wind fungus, black bovine liver, milk milk fungus, blue head fungus, dry mushroom, chanterelle mushroom, matsutake, red chisel, white boletus, red mushroom, yellow raisin, chicken fir, old man's head , coral fungi, shiitake mushrooms, tea tree mushrooms, pleurotus eryngii, Tricholoma eryngii as templates, using primers T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com