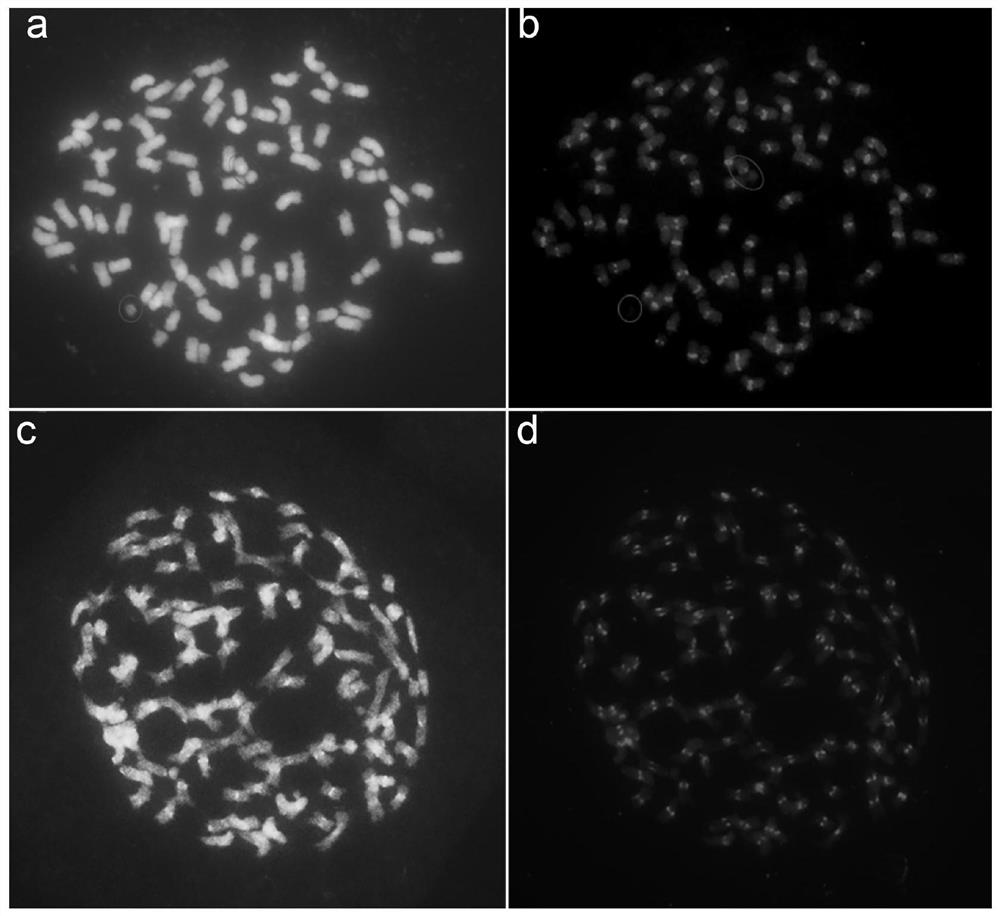
Novel method for carrying out accurate chromosome counting by applying saccharum centromere specific repetitive sequence
A repeat sequence, centromere technology, applied in the fields of bioinformatics and molecular cytogenetics, can solve problems such as difficulty in obtaining centromere sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Implementation of ChIP-Seq Technology
[0032] ChIP-Seq technology refers to the method of high-throughput sequencing of DNA obtained after chromatin immunoprecipitation ChIP.
[0033] The basic process of ChIP-Seq used in the present invention is:
[0034] 1. Grind fresh and young leaves with liquid nitrogen to extract cell nuclei;
[0035] 2. Extract the nucleus, use micrococcal ribozyme to enzymatically hydrolyze the nucleus, and randomly break the chromatin into fragments smaller than 300bp;
[0036] 3. Add the antibody of centromere-specific histone CENH3, and bind to the target protein-DNA complex; divide the nuclei after enzymatic hydrolysis into three parts, one part is the internal reference (input), and one part is added to the pre-immune serum ( Mock), another CENH3-specific antibody was added to immunoprecipitate the protein-DNA complex, and inverted overnight at 4°C;
[0037] 4. Add Protein A to bind the antibody-target protein-DNA complex and...
Embodiment 2
[0050] Embodiment 2: Primer design and PCR amplification
[0051] Use the primer design software Primer Premier 5.0 to design primers for the centromere sequence CENTS. The set conditions are: first, the full-length or partial sequence of the nucleic acid sequence; second, the length of the primer is 21bp±5bp; third, the GC content of the primer sequence is 40-60%; the fourth is to avoid the formation of stable dimer or hairpin structure between the primers; the fifth is that the primers cannot be mismatched at the non-target site of the template sequence.
[0052] The conditions of PCR amplification were: pre-denaturation at 95°C for 3 min; denaturation at 95°C for 45 s, annealing at 57°C for 30 s, extension at 72°C for 90 s, 30 cycles, and final extension at 72°C for 7 min.
[0053] It should be noted that the band amplified by PCR should be a single band. The PCR product can be directly purified and recovered using the QIAGEN kit, the product number: 28104, name: QIAquick ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com