Triple TaqMan fluorescent quantitative PCR detection reagent, kit, detection method and application
A fluorescence quantitative and detection reagent technology, applied in the field of pathogen detection, to achieve the effect of low cost, high sensitivity and improved work efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] A triple TaqMan fluorescent quantitative PCR detection reagent, including specific qPCR detection primers for novel coronavirus (SARS-CoV-2), feline calicivirus (FCV), influenza A (influenza A virus, IAV) and The corresponding TaqMan probes are as follows:
[0045] (1) The primer sequence and TaqMan probe sequence used to detect SARS-CoV-2 are: upstream primer N-112F, downstream primer N-112R, TaqMan probe N-probe;
[0046] The N-112F has a sequence as shown in SEQ ID No.1
[0047] The N-112R has a sequence as shown in SEQ ID No.2;
[0048] The N-probe has a sequence as shown in SEQ ID No.3;
[0049] The 5' end of the probe N-probe is labeled with a reporter fluorescent dye of HEX, and the 3' end of which is marked with a fluorescent quencher group of BHQ1: 5'-HEX-TTCACCGCTCTCACTCAACAT-BHQ1-3'.
[0050] (2) The primer sequence and TaqMan probe sequence used to detect FCV are: upstream primer FCV-106F, downstream primer FCV-106R, TaqMan probe FCV-probe;
[0051] The ...
Embodiment 2
[0063] Based on the triple fluorescent quantitative qPCR detection method of the detection reagent described in Example 1, the specific steps are as follows:
[0064] (1) Obtain the cDNA of the sample to be tested: extract the total RNA of the cat nasopharyngeal swab sample or respiratory tissue or organ, and reverse transcribe to obtain the cDNA template.
[0065] The reverse transcription was carried out using HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper). The total reaction system was 20 μL, including 1 μL Random hexamers, 6 μL RNase-free Water, 5 μL total RNA, and heated at 65°C for 5 min after mixing, and quickly Place it on ice for quenching, and let it stand on ice for 2 minutes, then continue to add 4×gDNA wiperMix, 4 μL, then gently blow and mix with a pipette, 42°C for 2 minutes; finally add 2 μL 10×RT Mix, 2 μL HiScriptII Enzyme Mix , after mixing, 5min at 25°C, 45min at 50°C, and 2min at 85°C to get it.
[0066] (2) Three sets of qPCR primers and probes ...
Embodiment 3
[0071] The construction and verification of the reagent described in Example 1 and the detection method described in Example 2, the specific experimental process is as follows:
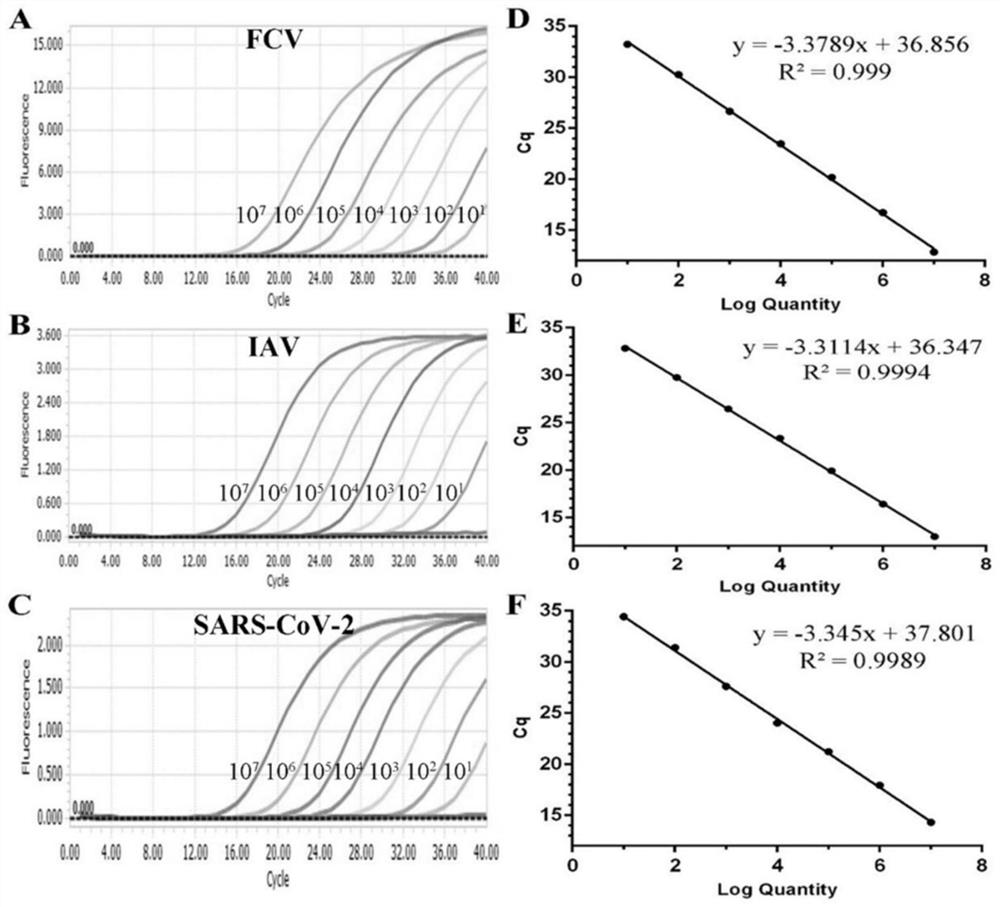
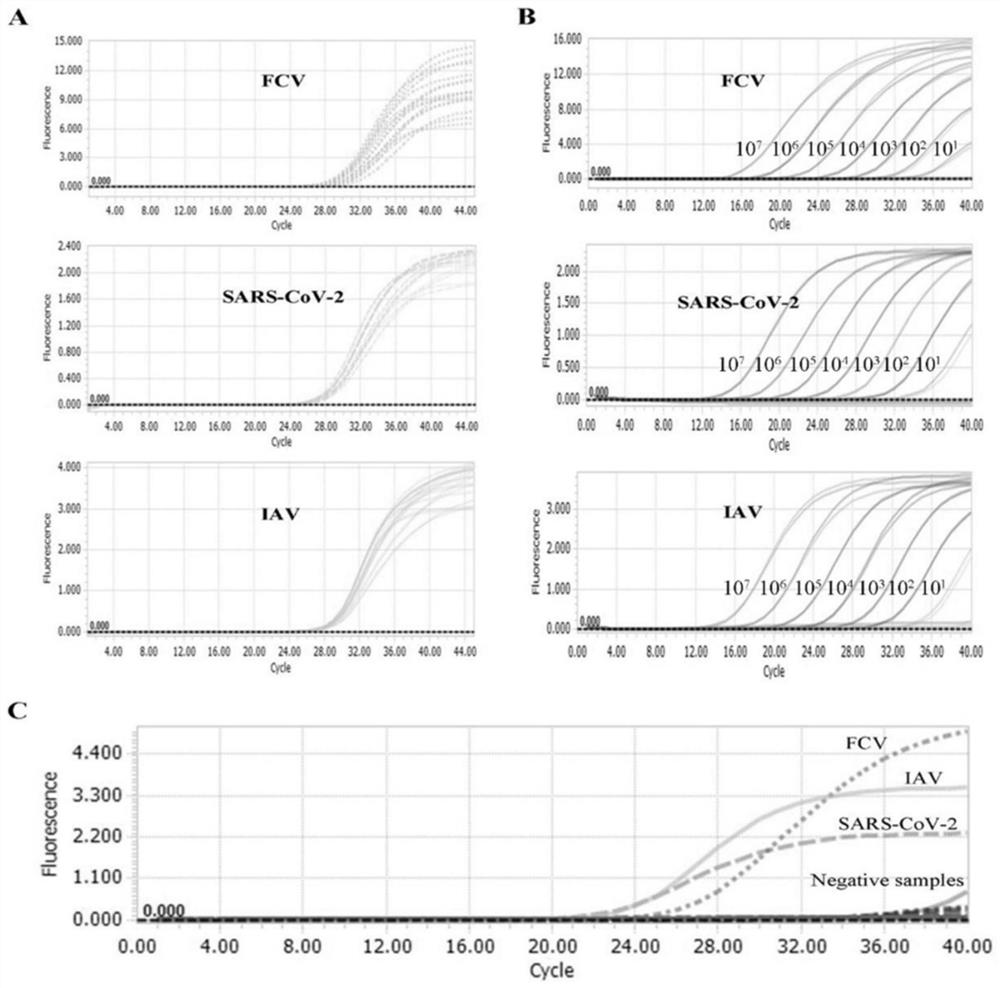
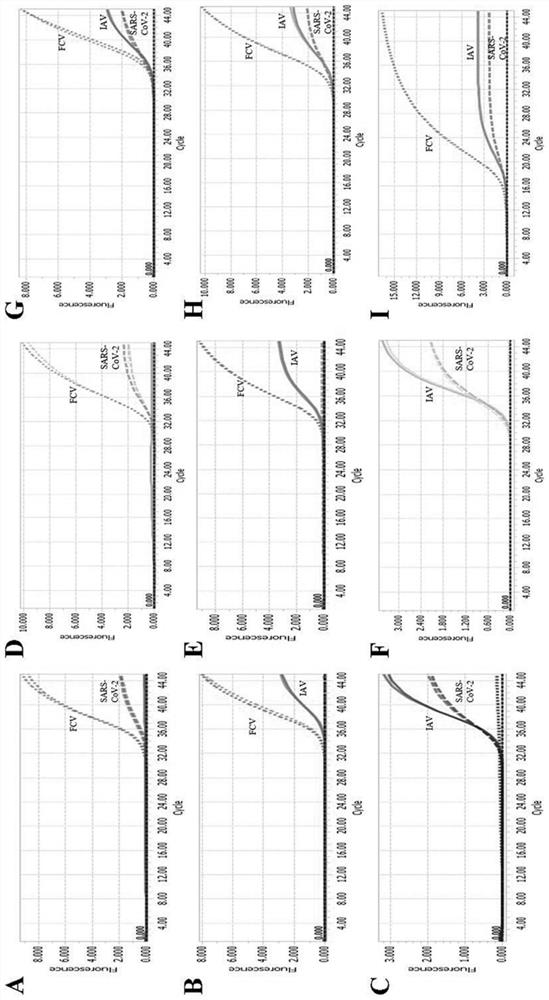
[0072] 1. Establishment of standard curve
[0073] (1) Design of primers and probes: All probes and primers in this experiment were designed using the primer design software Oligo 7, and were synthesized by Shanghai Sangon Bioengineering Co., Ltd.
[0074] (2) Preparation of plasmid standards: Using the primers N-112F (F / R), FCV-106F (F / R), and IAV-149F (F / R) designed in this experiment, the high-fidelity PCR enzyme Phanta Max Super- Fidelity DNA Polymerase (Nanjing Novizan Biotechnology Co., Ltd.) was used to amplify the target fragment, and then the target fragment was connected to the pMD18-T vector (Bao Bioengineering (Dalian) Co., Ltd.) vector to construct a recombinant plasmid. The reaction system and experimental program settings of the PCR amplification reaction were set according to the inst...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com