Engineered Cas12i nuclease as well as effect protein and application thereof
A nuclease, engineering technology, used in genetic engineering, hydrolase, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0320] Example 1: Replace the amino acid that interacts with PAM in the reference Cas12i2 enzyme with a positively charged amino acid, and verify its gene editing efficiency.
[0321] Plasmid construction
[0322] The coding sequence of Cas12i2 was codon optimized (human) and synthesized. Variants of the Cas protein were generated by PCR-based site-directed mutagenesis. The specific method is to divide the DNA sequence design of the Cas12i2 protein into two parts centered on the mutation site, design two pairs of primers to amplify the two parts of the DNA sequence, and introduce the sequence to be mutated into the primers, and finally clone by Gibson Both fragments were loaded into the pCAG-2A-eGFP vector. The combination of mutants is constructed by splitting the DNA of the Cas12i2 protein into multiple segments and using PCR and Gibson clone. The position of the mutant was determined by analyzing the structural information of Cas12i2 using protein structure visualization...
Embodiment 1-A
[0327] Example 1-A Four engineered Cas12i2 with single amino acid substitutions were selected
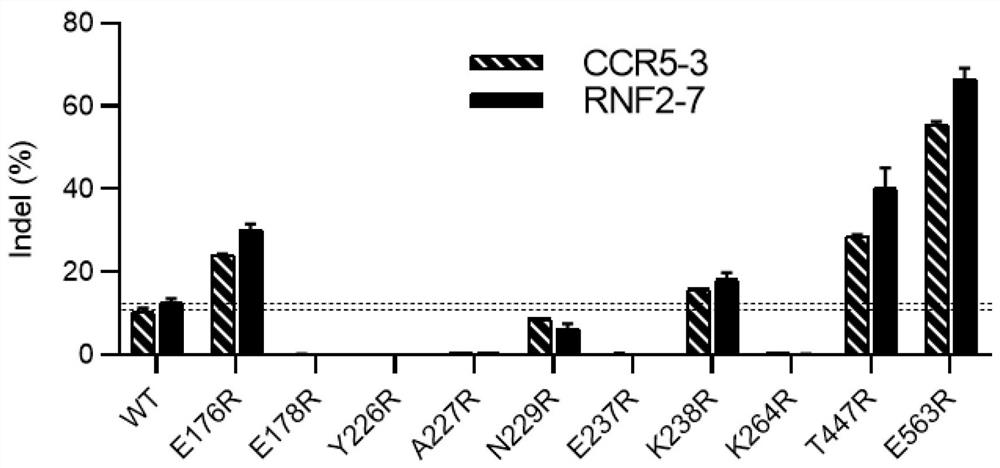
[0328]According to the method described in Example 1, engineered Cas12i2 enzymes with a single mutation in the amino acid sequence were respectively expressed, and the preferred amino acid replacement mode and its corresponding gene editing efficiency are shown in Figure 1 and Table 1. In Figure 1, we first selected 10 amino acids within 9 Å of Cas12i2 from PAM DNA: E176, E178, Y226, A227, N229, E237, K238, K264, T447, E563, and carried out point mutations of arginine (R) test. By comparing the gene editing efficiency of these mutants and wild-type Cas12i2 at two genomic loci: CCR5-3, RNF2-7 in 293T cells, we found that mutants with the following amino acid substitutions: E176R, K238R, T447R and E563R can effectively improve the gene editing efficiency (shown in Figure 1, there are four amino acid substitutions obtained higher than about 10% (this is the gene editing efficiency of ...
Embodiment 1-B
[0329] Example 1-B compares the engineered Cas12i2 with multiple preferred amino acid substitutions
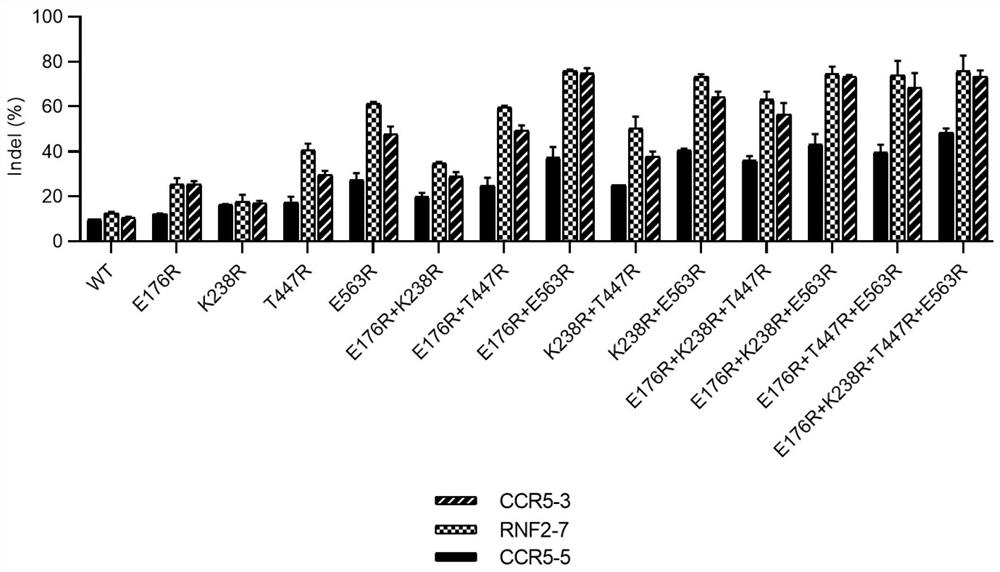
[0330] According to the method described in Example 1, respectively express the engineered Cas12i2 enzyme whose amino acid sequence has more than two preferred amino acid substitutions, the combination mode and gene editing efficiency comparison are shown in Table 1 and figure 2 . figure 2 It is shown that we combined the point mutations in the four efficiency-improving mutants, E176R, K238R, T447R and E563R, which were screened in Example 1-A. By comparing the gene editing efficiency of these mutants and wild-type Cas12i2 at three genomic loci: CCR5-3, CCR5-5, and RNF2-7 in 293T cells, we found that the efficiency can be further improved after the combination of point mutations mutant. Especially when the four mutations are combined (E176R+K238R+T447R+E563R), the most efficient combined mutation can be obtained.
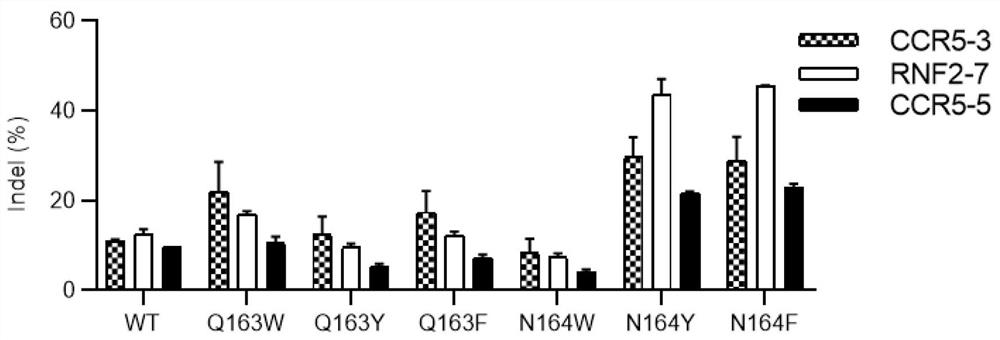
[0331] The experimental result summary of embodiment 1
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com