Method for detecting nucleic acid molecules in exosomes
A detection method and technology of nucleic acid molecules, applied in the field of detection of RNA and DNA contained in exosomes, can solve problems such as unfavorable extraction and quantification of RNA or DNA, loss of RNA or DNA molecules, influence analysis of results, etc., so as to overcome mutual inhibition. , Improve detection sensitivity and save consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
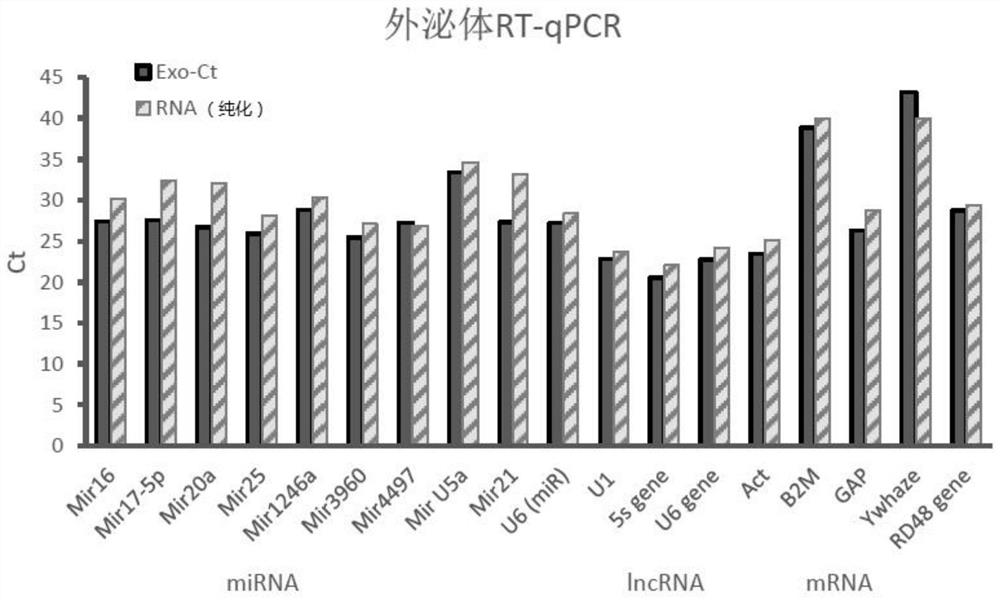
[0067] Example 1: Simple and rapid detection of exosomal RNA in Hek293 cell culture medium by one-step method
[0068] In this example, the recovery efficiency of exosome sample RNA was mainly compared using different methods. The exosome direct lysis (one-step) method of the present invention (ie, A, as the experimental group) was compared with the conventional RNA extraction method (multi-step) method (ie, B, as the control group). For the convenience of comparison, in the experimental group and the control group, except that the exosome RNA sample extraction steps are different (respectively direct lysis method and conventional RNA extraction method), other operating steps are the same, for example, both use the method of the present invention RT-For-All reverse transcription reaction for exosome nucleic acid detection. The specific operation is as follows:
[0069] 1. Experimental materials
[0070] (1) Main equipment: centrifuge (Thermo), PCR instrument (Takara), real-...
Embodiment 2
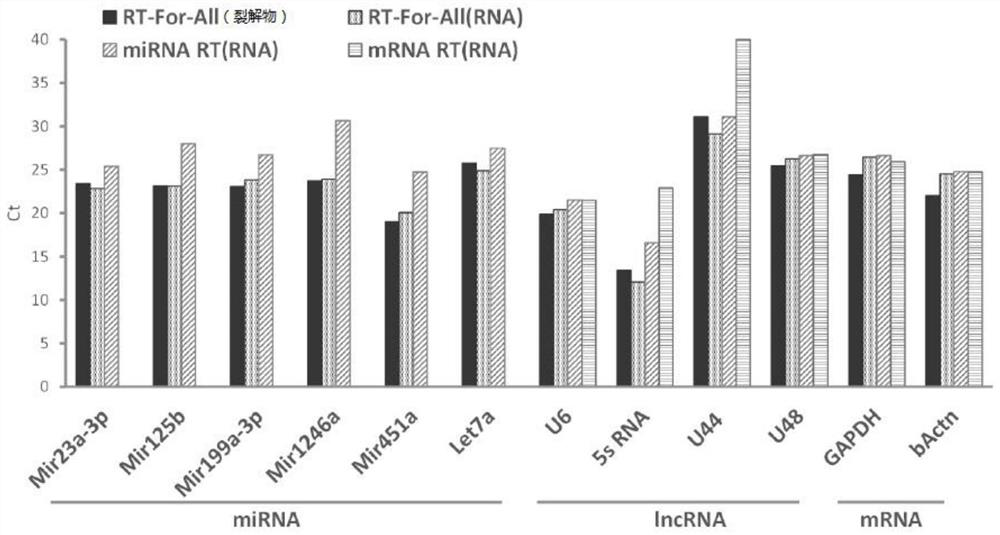
[0101] Example 2: One-step method for simple and rapid detection of exosomal RNA in mesenchymal stem cell (MSC) culture medium
[0102] In this example, the method of the present invention is completely applied to the detection of exosomes of mesenchymal stem cells (MSC). According to the latest literature reports, the marker miRNAs and housekeeping RNAs in the exosomes of mesenchymal stem cells (MSCs) were selected for detection, including Mir23a-3p, Mir125b, Mir1246a, etc.
[0103] In this example, in the reverse transcription step of the exosome RNA detection method, the RT-For-All reverse transcription reaction (ie, A, experimental group) in the method of the present invention was combined with the conventional miRNA reverse transcription reaction (ie, B, Control 1) was compared with a conventional mRNA reverse transcription reaction (ie, C, Control 2).
[0104] Among them, the conventional mRNA reverse transcription reaction (i.e., control group 2) can only detect some m...
Embodiment 3
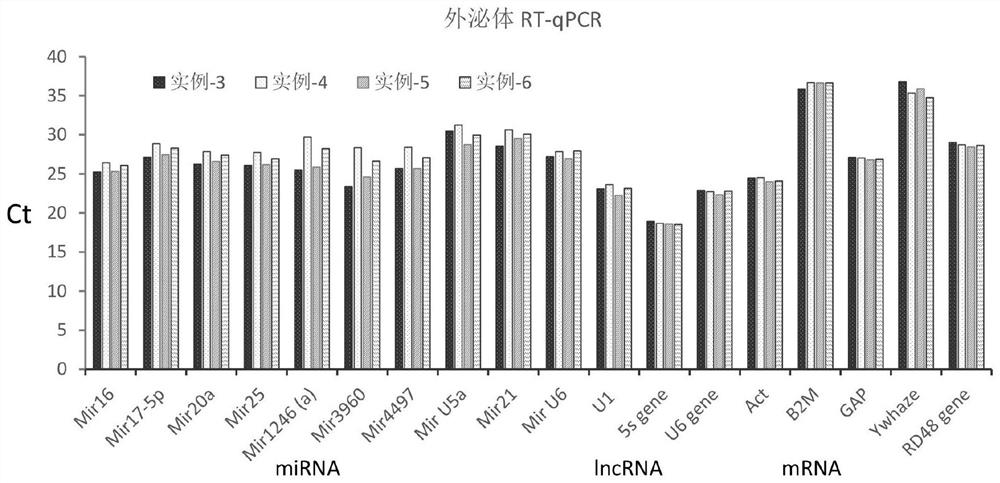
[0124] The operation method of Example 3 is similar to that of the experimental group in Example 1. The sample is ExoCT exosome lysate obtained by lysing exosomes with ExoCt lysis buffer. The difference is that reverse transcriptase and PolyA polymerase in the reverse transcription reaction system The concentrations of , miRNA Adapter and random primers are different, as shown in Table 1. In the reverse transcription reaction system, the concentration of reverse transcriptase is 1U / μL reaction system, and the concentration of the PolyA polymerase is 0.05U / μL reaction system; the concentration of miRNA Adapter in the RT buffer solution in the reverse transcription reaction is 0.2 μM, and the concentration of random primers is 0.2 μM. Test results image 3 As shown, it is found that it can achieve similar technical effects.
[0125] Table 1 embodiment 3-6 reverse transcription reaction system component concentration
[0126] RT-For-ALL Example 3 Example 4 Exam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com