EBER probe for detecting EBV infected tissue and detection kit
A technology for detection kits and probes, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, and methods based on microorganisms. It can solve the problems of cumbersome processes, poor solubility of PNA, and high synthesis costs, and achieve accurate and good detection results. The effect of hybridization specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
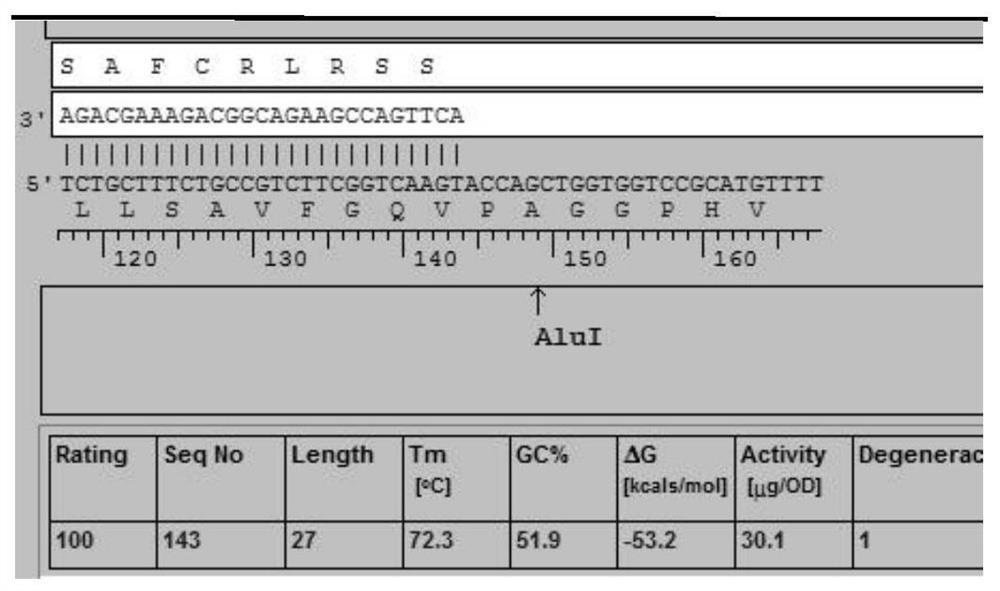
[0052] EBER probe design and synthesis
[0053] The EBER-1 and EBER-2 genes are EBV-encoded small RNAs that bind to host proteins and do not encode proteins, but are abundantly expressed in all EBV-positive cells, up to 10 in each cell 6 The copy can be used as the target of ISH to detect EBV. Retrieve the sequence (Human herpesvirus 4isolate SNU-1103EBER-1and EBER-2genes, completesequence, GenBank: EF187853.1), and analyze the sequence specificity, select the appropriate segment to design the EBER probe (see figure 1), to obtain the EBER-2 probe (5'-FAM-acttgaccgaagacggcagaaagcaga-3', SEQ ID NO: 1). At the same time, a control probe was synthesized with reference to the literature (Chinese patent application number CN201811313293.3), named EBER-1 probe (5'-FAM-ctcctccctagcaaaaccctcaggacggcg-3', SEQ ID NO: 2), used to compare new Set the performance of the probe.
Embodiment 2
[0055] Evaluation of two probes and two hybridization buffers
[0056] The two probes were formulated with two hybridization buffers to form a hybridization system, and the confirmed EBV-positive and negative nasopharyngeal carcinoma tissue samples FFPE sections were used for testing.
[0057] 1. System configuration
[0058] System 1 contained 10% dextran sulfate, 10mM NaCl, 30% formamide, 0.1% sodium pyrophosphate, 0.2% polyvinylpyrrolidone, 0.2% Ficoll, 50mM Tris-HCl (pH 7.5) and a final concentration of 5 μM fluorescently labeled probe needle (EBER-1 probe or EBER-2 probe).
[0059] System 2 contained 10% dextran sulfate, 50% formamide, 1% Triton X-100, 50mM Tris-HCl (pH 7.5) and a final concentration of 5 μM fluorescently labeled probe (EBER-1 probe or EBER-2 probe Needle).
[0060] 2. Detection method:
[0061] 1. Pre-treatment process:
[0062] Bake slices: bake slices at 65°C for 30 minutes.
[0063] Dewaxing: Environmentally friendly dewaxing agent twice for 10 ...
Embodiment 3
[0077] Method for detecting samples with FITC-labeled EBER-2 probe
[0078] 1. System preparation of probe hybridization solution
[0079] The system contained 10% dextran sulfate, 50% formamide, 1% Triton X-100, 50 mM Tris-HCl (pH 7.5) and a final concentration of 5 μM FITC-labeled EBER-2 probe.
[0080] 2. Detection method
[0081] The same method as in Example 2, wherein the sample numbers are 98172, K57084, K56035, K56302, K56926, K57085 and K55290 respectively.
[0082] 3. Test results
[0083] The results are shown in Figure 4.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com