Aptamer of methotrexate, aptamer derivative and application of aptamer and aptamer derivative
A technology of nucleic acid aptamer and methotrexate, applied in biological testing, biochemical equipment and methods, instruments, etc., can solve the problems of large toxic and side effects, narrow therapeutic window, endangering the life safety of patients, etc., and achieve small molecular weight, The effect of small batch variance and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1: Methotrexate nucleic acid aptamer screening.
[0037] 1. Synthesize a random single-stranded DNA library (GGAGGCTCTCGGGACGACNNNNNNNNNNNNNNNNNNNNNNNNNNNNGTCGTCCCGATGCTGCAATCGTAA), a biotin-labeled complementary strand to the single-stranded DNA library (TCCCGAGAGCCTCCTTTT-Biotin), a forward primer (GGAGGCTCTCGGGACGAC) and a biotin-labeled reverse primer (GGCAGACGATT)
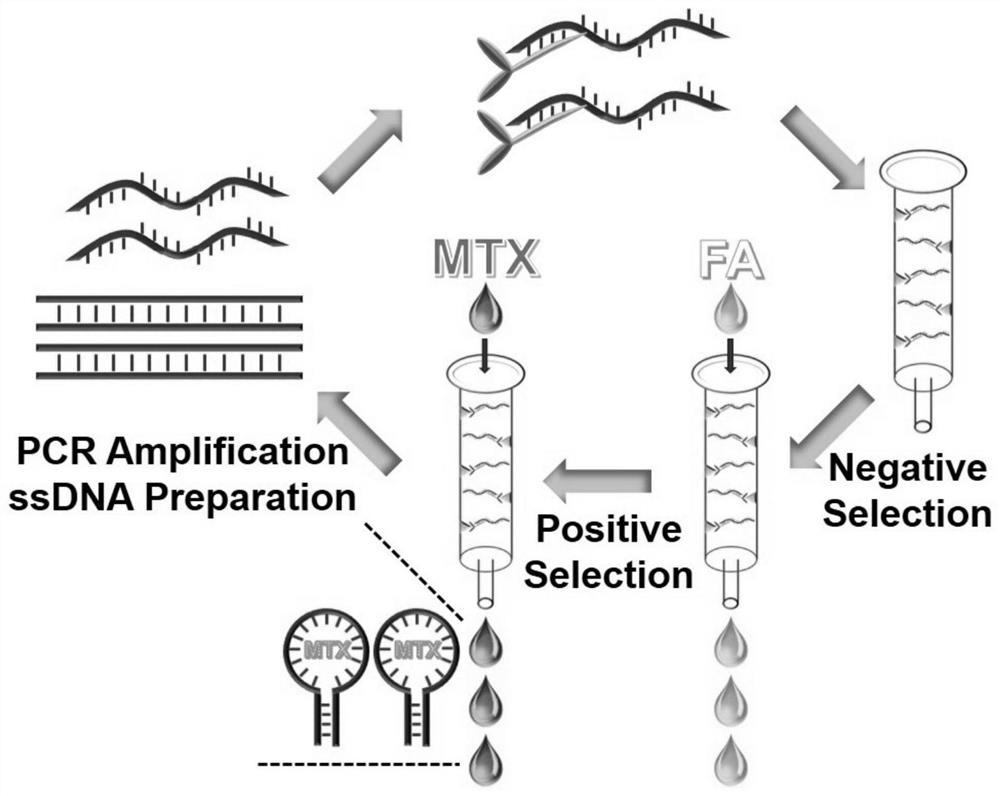
[0038] 2. Detailed screening process ( figure 1 )as follows:
[0039] Step 1: Take 1 nmol of ssDNA library strand and 5 nmol of complementary strand in 250 μL PBS (0.1M) buffer solution at 95°C for 5 min and then place at room temperature for 1 h;
[0040] Step 2: Take 200 μL streptavidin agarose balls to the micro-separation column, wash the streptavidin agarose balls in the separation column three times with PBS (200 μL / time), and then heat the denatured mixture Add to a miniature separation column and incubate with streptavidin agarose balls at room temperature for 10 min, collect the efflu...
Embodiment 2
[0053] Example 2: Construction of strand displacement biosensors.
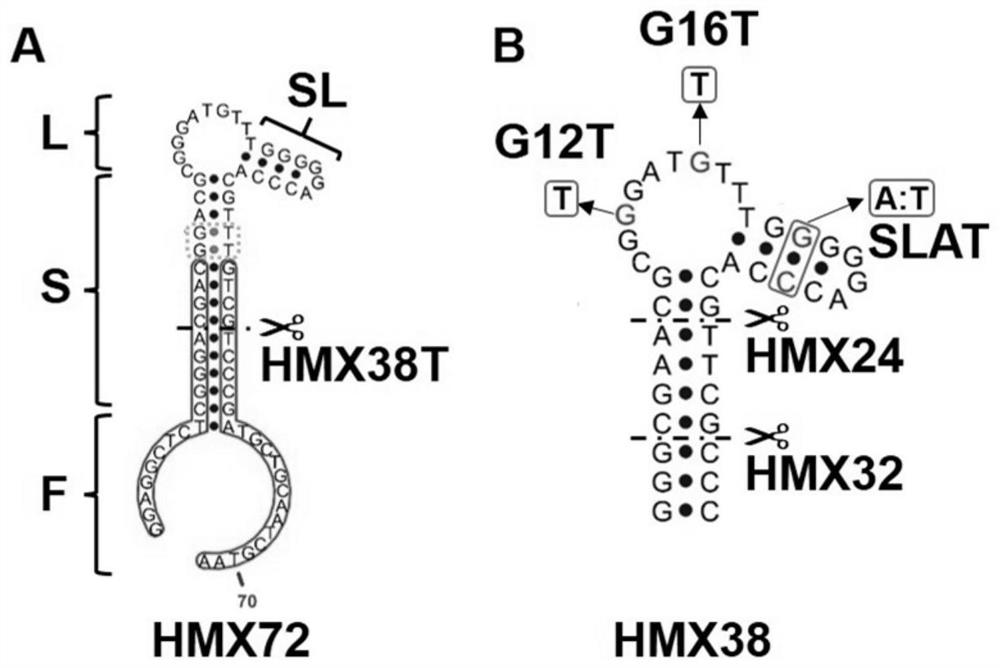
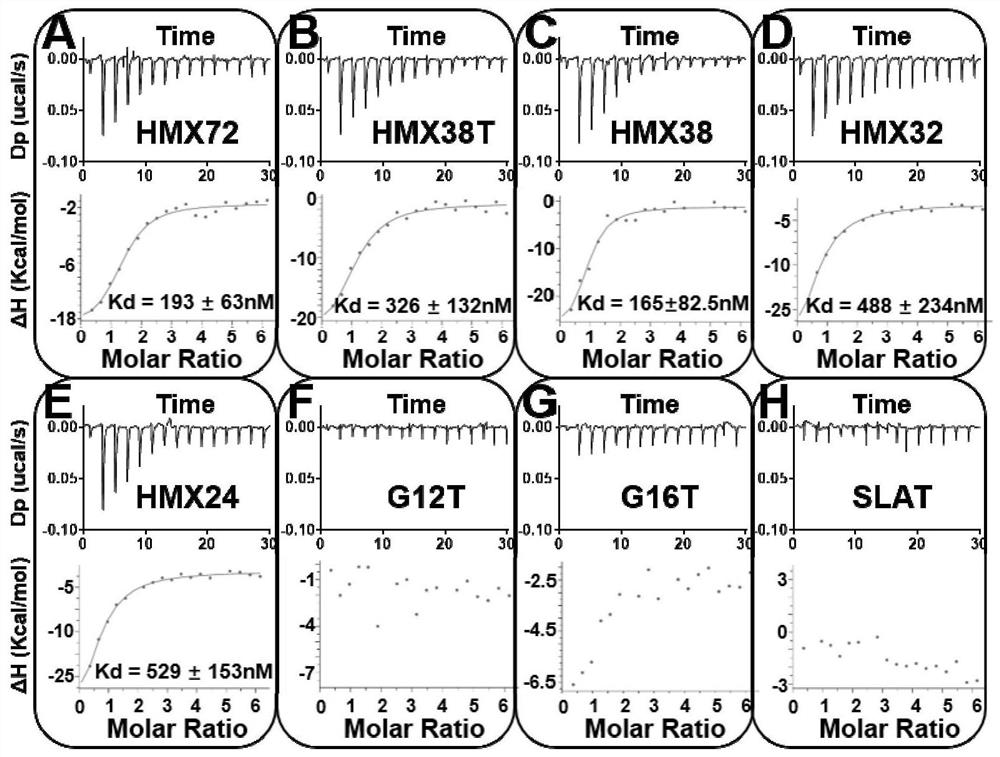
[0054] The DNA sequence of FAM-HMX38 (FAM-CTCTCGGGCGAACGCGGGATGTTTGGGGGACCCACGTTCGCCC) tagged with FAM at the 5' end and cap38-BHQ1 (CGTTCGCCCGAGAG-BHQ1) tagged with BHQ1 at the 3' end was synthesized.
[0055] The synthesized DNA powders were all dissolved in PBS and quantified to 10 μM by Nanodrop. Take 10 μL of FAM-HMX38 (final concentration of 2 μM), 30 μL of cap38-BHQ1 (final concentration of 6 μM) and 10 μL of PBS, heat at 95 °C for 5 min, and then place at room temperature for 1 h. cap38-BHQ1 can complementarily pair with the 5' end of FAM-HMX38 to form strand displacement biosensor 38-3 (final concentration 2 μM, this concentration is based on FAM-HMX38), such as Figure 4 . Add different concentrations of MTX (0 μM, 0.1 μM, 0.2 μM, 0.5 μM, 1 μM, 2 μM) to the above sensor 38-3 (final concentration 0.1 μM), react at room temperature for 30 min, and measure with a microplate reader. Its fluorescence v...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com