Economical and practical nucleic acid chain 5'-hydroxyl phosphorylation method
A technology of hydroxyphosphoric acid and nucleic acid chain, applied in the field of biochemistry, can solve problems such as high price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
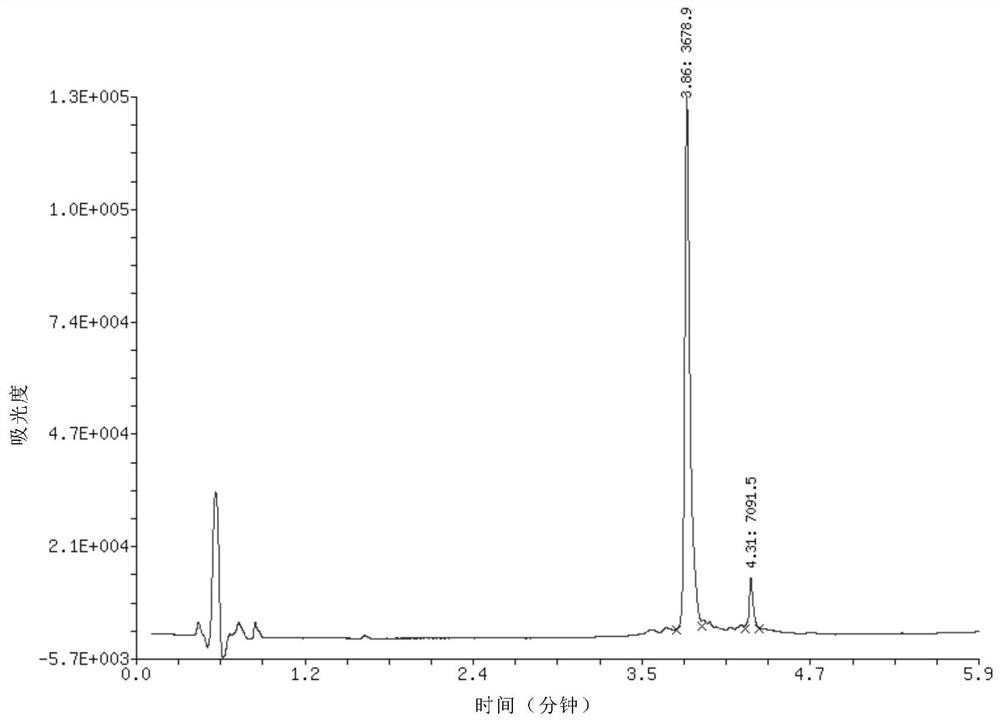
[0046] Example 1, 20 nanomoles of DNA single strand 3 before phosphorylation, with a length of 17 bases, a base sequence of CAGTGATGAGTCGGTCG, and a relative molecular mass of 5266 before phosphorylation, dissolved in nuclease-free water, and added to 20 microliters 10-fold phosphorylation buffer components, 5 units of T4 polynucleotide kinase, added nuclease-free water to a reaction volume of 50 microliters, and reacted at 37°C for 12 hours to complete the reaction. Add 5 microliters of 5 mol / liter sodium chloride aqueous solution and 125 microliters of absolute ethanol to the reaction solution, shake and mix, place in a -80°C refrigerator for 10-30 minutes, and centrifuge at a high speed (4°C, 12,000 rpm) / min, 5 minutes), a precipitate was obtained. The precipitate was dissolved in 20 microliters of water, and quantitatively detected by DNA ultraviolet spectrophotometry, the amount of the phosphorylated product was measured to be 17.5 nmoles, and the recovery rate was 88%.
Embodiment 2
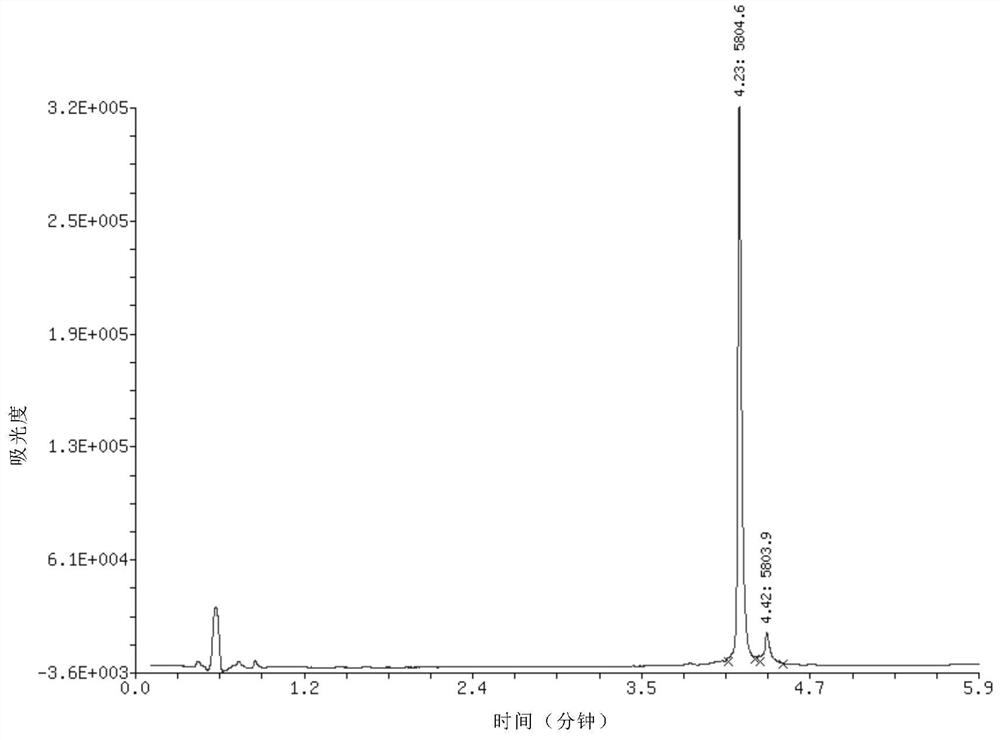
[0047] Example 2, 20 nanomoles of DNA single-strand 3 before phosphorylation, 17 bases in length, base sequence CAGTGATGAGTCGGTCG, relative molecular mass before phosphorylation is 5266, dissolved in nuclease-free water, and added to 20 microliters 10-fold phosphorylation buffer composition, 5 units of T4 polynucleotide kinase, 0.2 microliter of PEG, added nuclease-free water to a reaction volume of 50 microliters, and reacted at 37°C for 2.5 hours to complete the reaction. Add 5 microliters of 5 mol / liter sodium chloride aqueous solution and 125 microliters of absolute ethanol to the reaction solution, shake and mix, place in a -80°C refrigerator for 10-30 minutes, and centrifuge at a high speed (4°C, 12,000 rpm) / min, 5 minutes), a precipitate was obtained. The precipitate was dissolved in 20 microliters of water, and quantitatively detected by DNA ultraviolet spectrophotometry, the amount of the phosphorylated product was measured to be 17.4 nmoles, and the recovery rate wa...
Embodiment 3
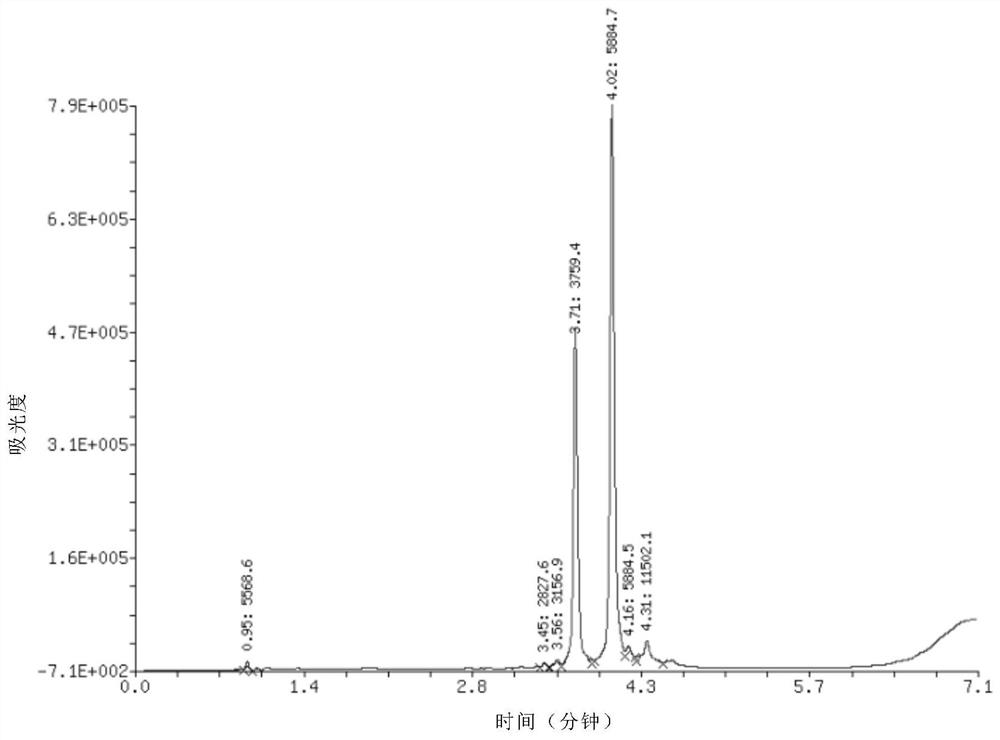
[0048] Example 3, 20 nanomoles of DNA single strand 3 before phosphorylation, with a length of 17 bases, a base sequence of CAGTGATGAGTCGGTCG, and a relative molecular mass of 5266 before phosphorylation, dissolved in nuclease-free water, and added to 20 microliters 10-fold phosphorylation buffer components, 5 units of T4 polynucleotide kinase, 0.5 microliter PEG, added nuclease-free water to make the reaction system 50 microliters, and reacted at 37°C for 2 hours to complete the reaction. Add 5 microliters of 5 mol / liter sodium chloride aqueous solution and 125 microliters of absolute ethanol to the reaction solution, shake and mix, place in a -80°C refrigerator for 10-30 minutes, and centrifuge at a high speed (4°C, 12,000 rpm) / min, 5 minutes), a precipitate was obtained. The precipitate was dissolved in 20 microliters of water, and quantitatively detected by DNA ultraviolet spectrophotometry, the amount of the phosphorylated product was measured to be 17.2 nmoles, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com