Hairpin-like adapter, construct and method for characterizing double-stranded target polynucleotide
A target polynucleotide, double-stranded nucleic acid technology, applied in the field of biological analysis and detection, can solve the problems of increasing the complexity of library preparation, increasing the complexity of preparation process, reducing the efficiency of library preparation, etc., and achieves high-reliability orthogonal correction reading ability, improved sequencing accuracy, and low synthesis difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
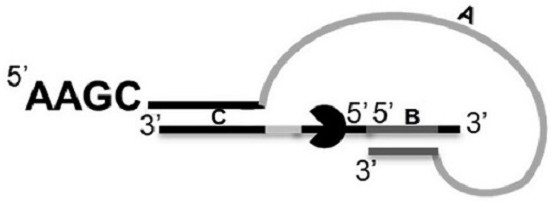
[0208] Example 1: Preparation of hairpin-like adapters
[0209] (1) 3'-5'-5'-3'( figure 1 From right to left in ) the synthesis of the second single strand
[0210] Will 3'-5' ( figure 1 From right to left in the first segment (SEQ ID NO: 2, 5' end has Azide (marked with N3) modification) and 5'-3' ( figure 1 From right to left in ) the second segment (SEQ ID NO:3, 5' end is modified with DBCO (marked by DB)) in a solution of 10mM Tris, 50mM NaCl, 0.1mM EDTA at a ratio of 3:1, react A click chemical reaction occurred at a temperature of 30° C. for 12 hours, and the obtained product was purified by PAGE to obtain a 3′-5′-5′-3′ second single chain.
[0211] (2) Synthesis of DNA containing a hairpin-like structure
[0212] The 5'-3' first single strand (SEQ ID NO: 1) and the 3'-5'-5'-3' second single strand formed in step (1) were lowered from 95°C by 0.1°C every 10s. Annealed at 4°C to form a hairpin-like structure DNA, such as figure 1 shown.
[0213] (3) Preparation of ...
Embodiment 2
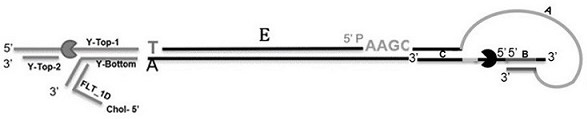
[0226] Example 2: Preparation of constructs
[0227] (1) Preparation of Y adapter
[0228] Y-Top-1 (SEQ ID NO:7), Y-Top-2 (SEQ ID NO:8) and Y-Bottom (SEQ ID NO:9) were annealed under the condition of decreasing 0.1°C every 10s from 95°C to 4°C to form the Y adapter.
[0229] (2) Preparation of enzyme-loaded Y adapter
[0230] The T4 Dda helicase was loaded onto the Y adapter by the method of steps (2)-(3) of Example 1.
[0231] (3) Preparation of constructs
[0232] An approximately 2700 base pair segment of plasmid DNA (SEQ ID NO: 6) was amplified using PCR primers containing restriction sites as defined below for the preparation of constructs containing the double-stranded target polynucleotide sequence.
[0233] BsaI forward primer: 5'-TCGCC ATTCA GGCTG CGC-3' (SEQ ID NO: 11)
[0234] BsaI reverse primer: 5'-GCTTA GAGAC CTGTG CGG-3' (SEQ ID NO: 12)
[0235] The fragment of about 2700bp was successively digested with a dA tail (NEB Cat#E7442) and BsaI restriction endon...
Embodiment 3
[0242] Example 3: Methods for Sequencing Double-Stranded Target Polynucleotides of Constructs Containing Hairpin-Like Adapters and Not Containing Hairpin-Like Adapters, respectively
[0243] Materials and methods:
[0244] Melt sequencing buffer (20mM HEPES pH 8.0, 50mM ATP, 50 mM MgCl 2 , 500 mMKCl), the first tether (SEQ ID NO:4, located at Figure 6 D) and the second tether (SEQ ID NO: 10 in image 3 FLT_1D on the left). Add 4ul of the above-mentioned tether (1uM) to 196ul of the sequencing buffer and mix well, then add it to the QNome-9604 sequencer of Qitan Technology and incubate for 20 minutes. The structure of the sequencer QNome-9604 is as follows Figure 12 As shown, it includes fluidic chip, signal processing circuit and sequencing software. Then, prepare the loading library, and the components are shown in Table 1 and Table 2.
[0245] Table 1: Preparation of Construct Sequencing Libraries Containing Hairpin-Like Adapters
[0246]
[0247] Table 2: Prepar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com