Protein for regulating pressure stress resistance of cronobacter sakazakii, and coding gene and application thereof
A technology with strong pressure resistance and bacilli, which is applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve problems such as no related reports, achieve broad application prospects, reduce integrity, and achieve good killing effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Discovery of Differential Expression of CpxA Gene of Cronobacter sakazakii
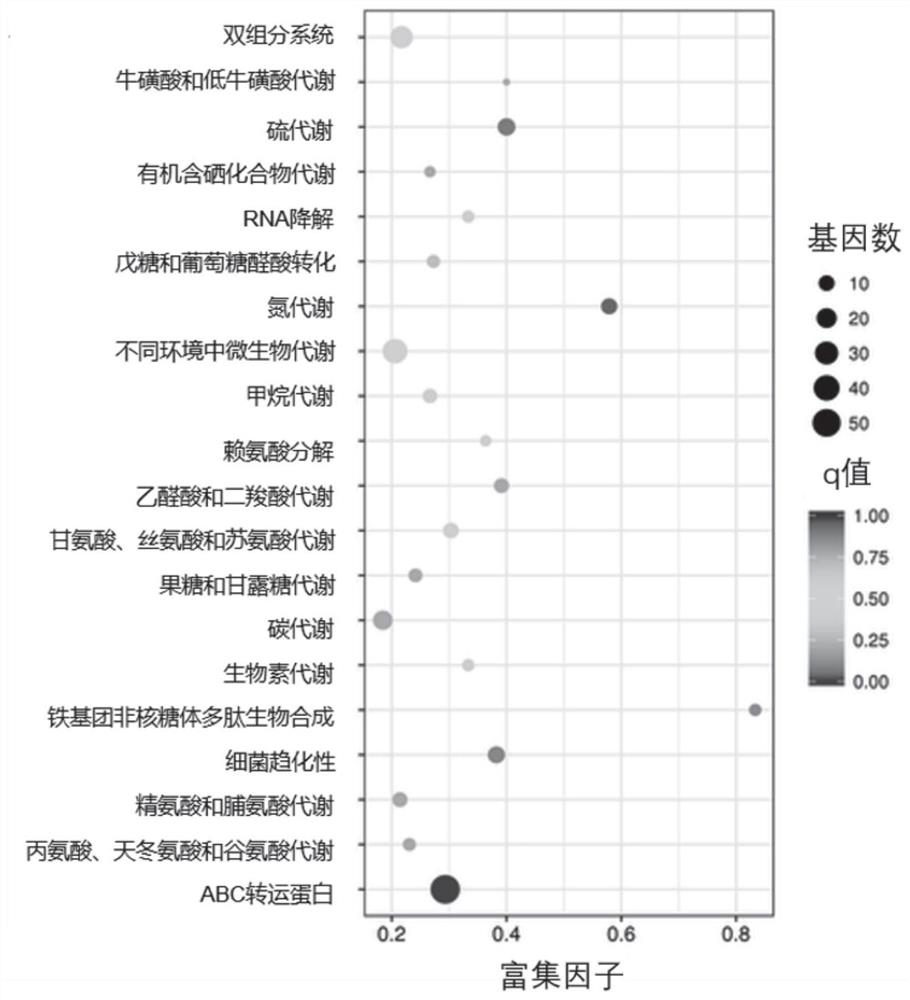
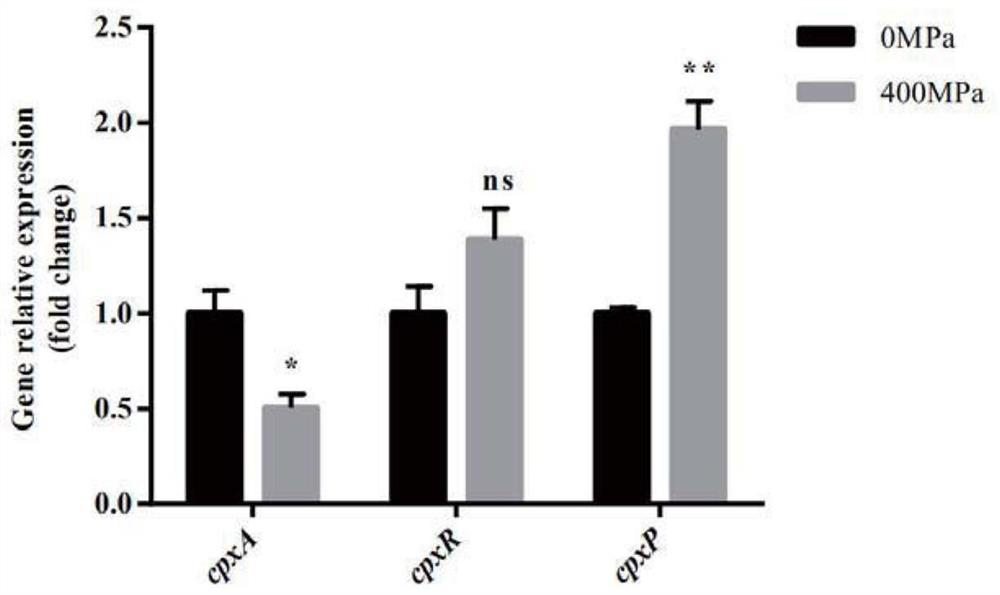
[0042]5mL of adjusted concentration of Cronobacter sakazakii suspension (about 10 8 CFU / mL) into 10mL polyethylene plastic bags, packaged and sealed, and subjected to 10min ultra-high pressure treatment under the conditions of 50MPa and 400MPa. The statistical enrichment of differentially expressed genes in the KEGG pathway was then analyzed using KOBAS software. It was found that the two-component system (Two-componentsystem) was differentially expressed after 50MPa treatment, see Figure 1A . The q-PCR verification of the differentially expressed genes of Cronobacter sakazakii under 400MPa ultra-high pressure conditions shows that under the condition of 400Mpa, the cpxA gene expressing histidine kinase in the Cpx two-component regulatory system of Cronobacter sakazakii Transcript levels are downregulated, see Figure 1B , indicating that under high-pressure environmental stress, ...
Embodiment 2
[0043] Cloning of embodiment 2 Cronobacter sakazakii CpxA gene
[0044] The genomic DNA of Cronobacter sakazakii was extracted as a template, and the following primers were designed according to the genomic sequence of the target gene:
[0045] The forward primer is cpxAF: AGGGCACGATGATTGAGCA
[0046] The reverse primer is cpxAR: CTGACCGATAAAGTTGCGAATG
[0047] After sequencing, the PCR amplification product of Cronobacter sakazakii CpxA has the nucleotide sequence shown in SEQ ID No:1.
Embodiment 3
[0048] Example 3 Construction of Cronobacter sakazakii mutant ΔCpxA
[0049] The following primers were designed according to the genomic DNA sequence of Cronobacter sakazakii:
[0050] ATCC-TF: GTCCCTGTTAAAGGAATTGCTCG
[0051] ATCC-TR: ATCAGCATTTCAACGGCATCA
[0052] ATCC-MF1: GGAATCTAGACCTTGAGTCGTTGCTCGACGTGATGATGCC
[0053] ATCC-MR1: GTGATAAAGCGGCAACCAGAGCCAGAAAATGGCGAAGATGC
[0054] ATCC-MF2: GCATCTTCGCCATTTTCTGGCTCTGGTTGCCGCTTTATCAC
[0055] ATCC-MR2: ACAGCTAGCGACGATATGTCTTTGTTGTTTCTGACGGTGGC
[0056] pLP-UF: GACACAGTTGTAACTGGTCCA
[0057] pLP-UR: CAGGAACACTTAACGGCTGAC
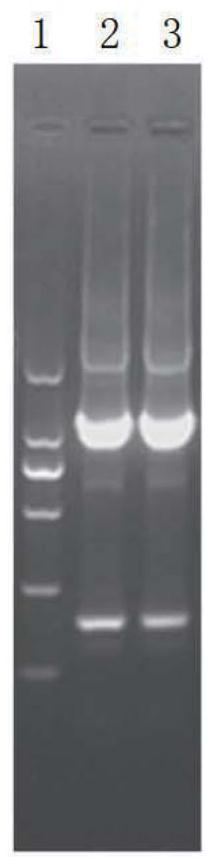
[0058] ATCC-MF1 / ATCC-MR1 and ATCC-MF2 / ATCC-MR2 were used to amplify, respectively, to obtain ATCC upstream homology arm A fragment and ATCC downstream homology arm B fragment. Then use the above A and B fragments as templates to perform overlapping PCR amplification to obtain the fusion AB fragment, see Figure 2A . The AB purified fragment was connected with the suicide vector pLP12cm, and the re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com