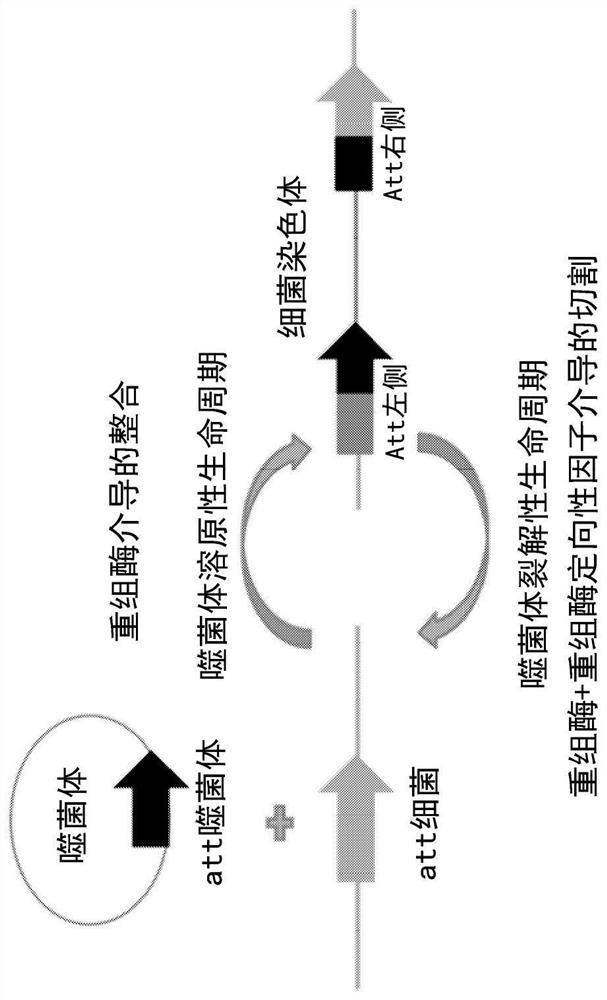
Serine recombinases mediating stable integration into plant genomes
A plant genome and recombinase technology, applied in genetic engineering, recombinant DNA technology, and stably introducing foreign DNA into chromosomes, can solve problems such as no sequence or structural similarity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0332] Example 1 Plasmid Construct
[0333] 1. Recombinase plasmid
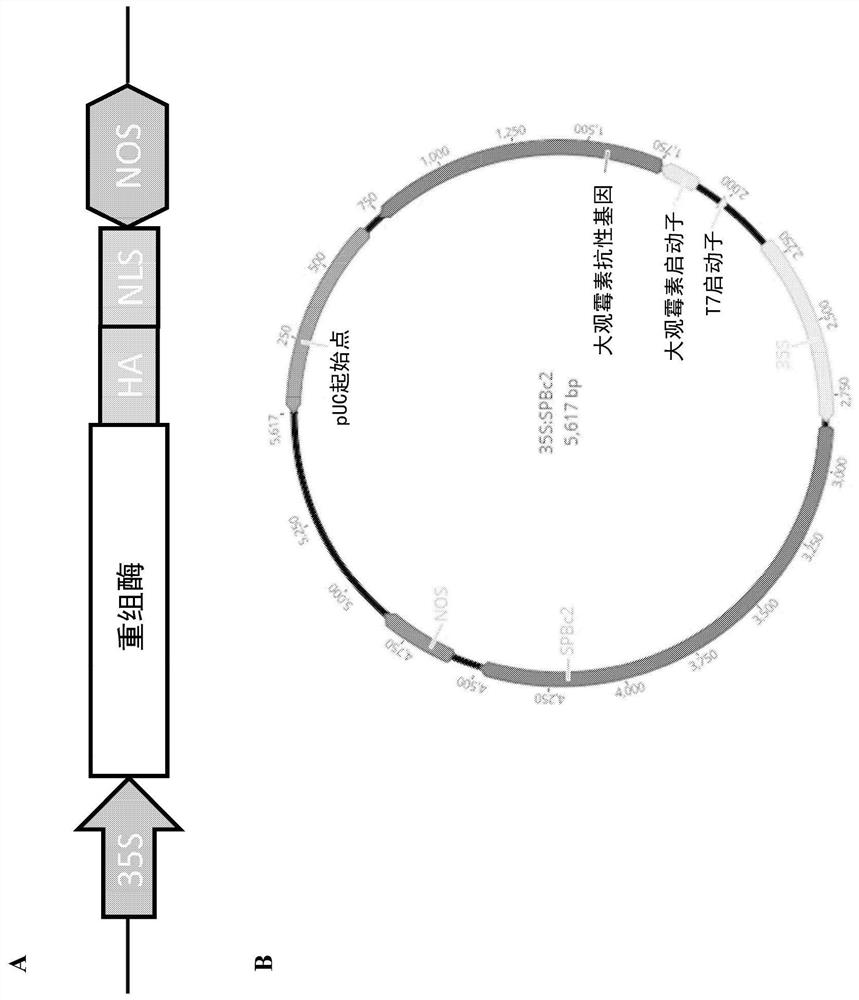
[0334] Plasmids with recombinases are based on the pUC backbone. Each recombinase gene was modified to express a fusion protein with a C-terminal HA (influenza hemagglutinin) peptide and an SV40 nuclear localization signal (NLS). It is expressed under the control of cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase (NOS) terminator. figure 2 is a schematic diagram of a recombinase construct expressing a recombinase fusion protein.
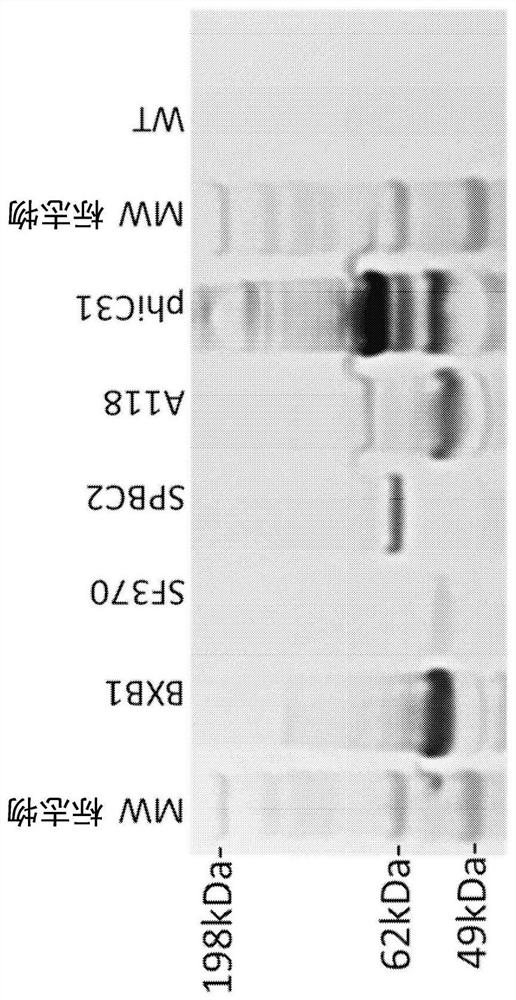
[0335] Table 2 below describes the phage as the original source of the recombinase, the structure of the recombinase fusion protein used in this study, and the amino acid sequence identifier.
[0336] Table 2: Sources of Recombinase
[0337]
[0338] In experiments without effector plasmids, to balance the amount of DNA transfected into cells, a plasmid expressing the chloramphenicol acetyltransferase (CAT) gene was used as a control for transfection (Ulma...
Embodiment 2
[0349] Example 2. Genbank screening of lettuce and tobacco genomes did not find attP or attB sites
[0350] Screening of attB, attP and possible pseudo att sites was performed against public genome databases. Lettuce (Lactuca sativa) and Nicotiana benthamiana genomes in the Phytozome database (from the US Energy Joint Genome Institute, available at Phytozome.jgi.doe.gov / pz / portal.html) were examined. The Arabidopsis genome was searched at Arabidopsis.org, a database established by Arabidopsis Information Sources (TAIR).
[0351] These databases were searched for the attB and attP sequences listed in Table 3, allowing for 2 nucleotide mismatches or spacers to account for possible att "pseudo" sites. The att pseudosites identified in US9034650 were also screened against lettuce and tobacco genomes.
[0352] No attP or attB sites or canonical pseudosites were found in Arabidopsis, lettuce, and tobacco. Therefore, serine recombinase cannot incorporate reporter plasmids with att...
Embodiment 3
[0353] Example 3.SPβc2 recombinase integrates the 6.3kb plasmid containing attP but not attB into the lettuce genome
[0354] 1. Method
[0355] E. coli containing effector or reporter plasmids, respectively, were grown and DNA was extracted using the Qiagen Endo-Free Maxi kit (Cat# 12361, Qiagen, Inc., CA).
[0356] Mesophyll protoplasts were isolated from 3-5 week-old axenically grown lettuce plants as described by Tiwari et al., 2006 (Tiwari et al., "Transfection assay with protoplasts containing an integrated reporter gene", Methods Mol Biol, 2006 ; 323:237-44). Purified protoplasts were transfected with a plasmid expressing the recombinase using the standard polyethylene glycol method as described by Tiwari 2006 (supra).
[0357] The protocol for the subculture and regeneration of transfected protoplasts was as previously described (Jie et al., "Myo-inositol increases the plating efficiency of protoplasts of cabbage (Brassica oleracea var. capitata) cotyledon derived f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com