Mutated motor protein, application of mutated motor protein and kit comprising mutated motor protein
A motor protein and domain technology, applied in the field of molecular biology, can solve the problems of rolling circle replication amplification application limitation, low amplification sensitivity, false positive amplification, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
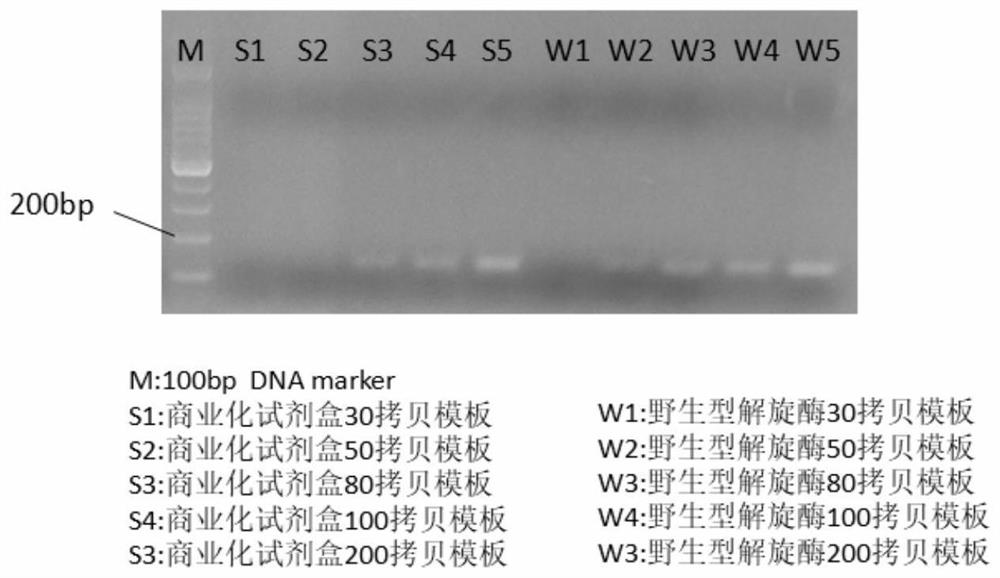
[0047] Application of commercial kits and wild-type Tte UvrD in isothermal amplification
[0048] The wild-type Tte UvrD DNA sequence is as follows:
[0049] SEQ ID No. 1:
[0050]
[0051]
[0052] The wild-type Tte UvrD amino acid sequence is as follows:
[0053] SEQ ID No.2:
[0054] MKEILANLNEQQKEAVTTTEGPLLILAGAGSGKTRVLTHRIAYLIKEKKVSPSNILAITFTNKAAEEMKTRVENLLGYVGDLWVSTFHSACVRILRRDIDKLGYDRNFVIFDTTDQKALVQECLKELDLSEKQYPIKMVLNAISSAKDKMVYPDDYIDFFGDTYRNRKIKEIYKLYQHKLKKINALDFDDIIIKTIELFKENPEILEFYQRKFRYIMVDEYQDTNTPQYYFVNLLAQRHRNLCVVGDDDQSIYGWRGADVRNILNFEKDYPEAKVIKLEQNYRSTKIILEAANHVIDNNVYRKKKSLWTQNKEGEKIVLCELENEREEAEFVIQEIIKLKERENRSFKDFAILYRTNAQSRPFEEALMKVKVPYKVVGALRFYDRKEIKDILAYLRLIVNPYDDISFKRIVNVPRRGIGPATIEALEKVAREKDTSLFFAIEDLKNARNKGSLLQFKQFILDLIDKKDAMSVSDLIKYILEQTGYIEELKREESEEAEGRIENLNEFLNAAYEFEESSEDKSLEAFLAGITLVSDIDMAGDIGESVVLMTLHSAKGLEFPVVFMVGMEEGLFPSYSSFEDDHELEEERRLCYVGITRSKERLYLTYARQRNLYGRSQYNSYSRFISEIPERLIVRYNIPTSKKTGFVSVHTFSDVYERSFSLGDKVEHKIWGIGTVVKVEGEEITVAFPNVGIKKL...
Embodiment 2
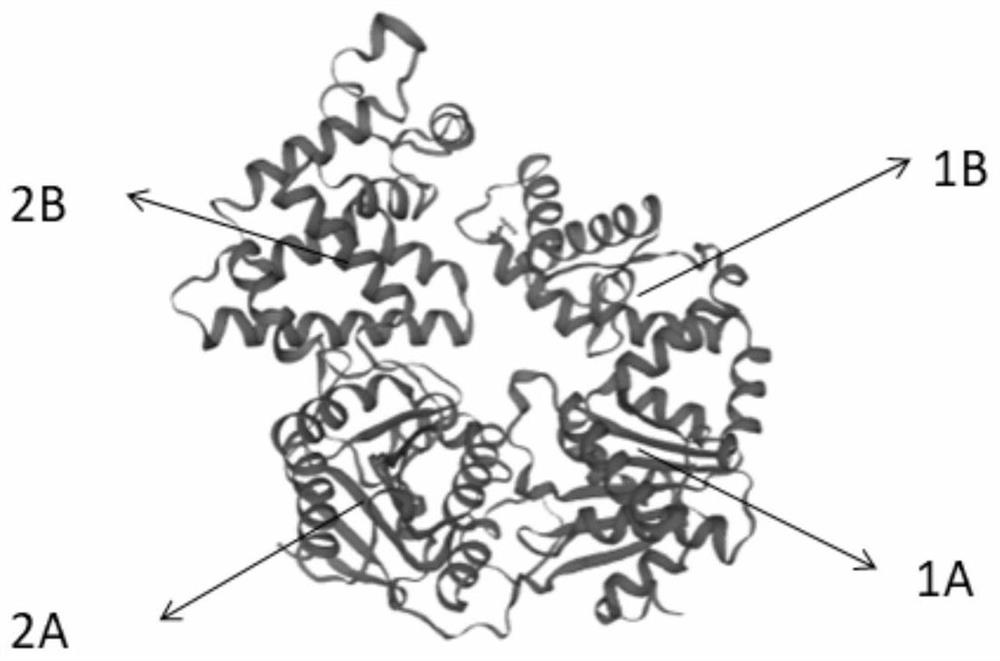
[0075] Effect of the I418G mutation in the 1B / 2B domain on the capture of double-stranded DNA by motor proteins
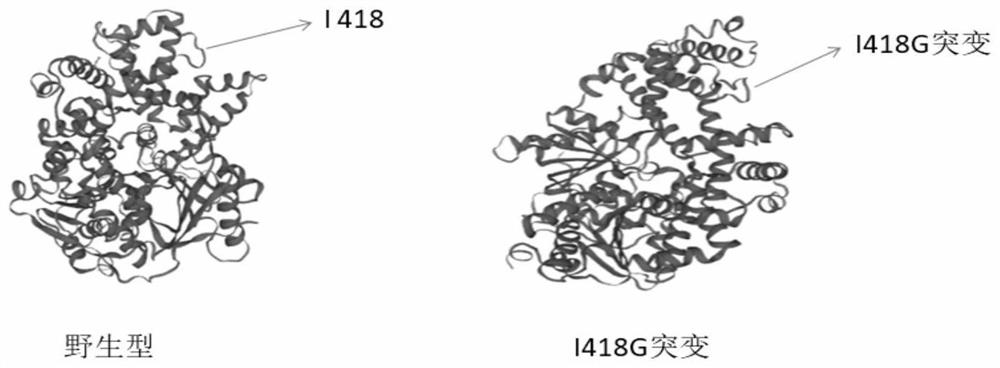
[0076] G417, I418, G419 and their surrounding regions constitute the circular part in the three-dimensional structure diagram, which is the key region for the helicase to bind and grab double-stranded DNA. image 3 A comparison of the three-dimensional structures of the wild-type enzyme and the I418G mutant enzyme is shown.
[0077] The expression experiment method of the I418G mutant enzyme is the same as the expression method of the wild-type enzyme in Example 1. The experimental system, target region, primers, etc. of the isothermal amplification reaction are also the same as in Example 1.
[0078] See the experimental results Figure 4 .
[0079] Example 2 Analysis of the results: Compared with the situation where the wild-type enzyme cannot amplify 30 copies of the template, the I418G mutant enzyme has a higher affinity for a small amount of DNA template (s...
Embodiment 3
[0081] Effects of P414K, R415I, R416I, G417E, I418L, I418R, I418T, I418V, P420V, A421K, T422D mutations in the 1B / 2B domain on the capture of double-stranded DNA by motor proteins
[0082] Figure 5 The three-dimensional structures of 11 individual point mutant enzymes are shown.
[0083] The expression method of the 11 kinds of single mutant enzymes is the same as the expression method of the wild-type enzyme in Example 1. The experimental system, target region, primers, etc. of the isothermal amplification reaction are also the same as in Example 1.
[0084] See the experimental results Figure 6 .
[0085] Example 3 Analysis of the results: Compared with the situation where 30 copies of the wild-type enzyme template cannot be amplified, the P414K mutation, R415I mutation, I418L mutation, and A421K mutation are due to the very small amount of DNA template (such as as low as 30 copies) in the system. There is a higher affinity grabbing ability, so it has been able to effe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com