Specific T cell epitope peptide P38 screened from novel coronavirus S protein whole proteome and application thereof
A coronavirus, specific technology, applied in the direction of viral peptides, applications, viruses, etc., to achieve the effect of good immune stimulation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Collection of blood samples from COVID-19 convalescent patients and unexposed population
[0034] Blood samples were collected from discharged patients with COVID-19 within two weeks after discharge, and PBMCs were immediately isolated and stored in liquid nitrogen. All COVID-19 patients are required to meet the discharge criteria for COVID-19 patients in the Diagnosis and Treatment Program for Novel Coronavirus Infected Pneumonia (7th Edition) promulgated by the General Office of the Chinese Health Commission. Blood PBMC samples from unexposed persons (no SARS-CoV-2 infection, no new crown vaccine) were collected as negative controls.
Embodiment 2
[0035] Example 2 SARS-CoV-2 Spike protein peptide library
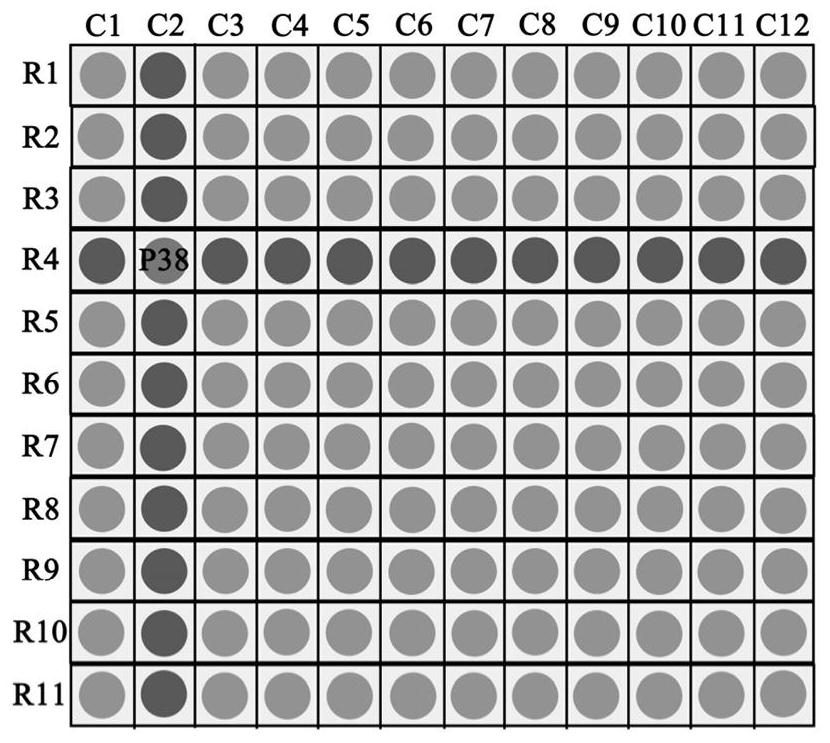
[0036] According to the amino acid sequence of the SARS-CoV-2 S protein (access number: YP_009724390.1), the length of the peptide was designed to be 15-mers, and two consecutive peptides overlapped by 5 amino acids. Finally, a total of 127 peptides covering the full length of the S protein were obtained (P1-P127), the amino acid sequence of the polypeptide is shown in Table 1. The 127 peptides were arranged in order into a checkerboard list of 12(C1-C12)×11(R1-R11), see Table 2, and 23 peptide groups were obtained.
[0037] Table 1 Peptide sequences of Spike protein peptide library
[0038]
[0039]
[0040] Table 2 Peptide checkerboard list
[0041] Peptide group C1 C2 C3 C4 C5 C6 C7 C8 C9 C10 C11 C12 R1 P1 P2 P3 P4 P5 P6 P7 P8 P9 P10 P11 P12 R2 P13 P14 P15 P16 P17 P18 P19 P20 P21 P22 P23 P24 R3 P25 P26 P27 P28 P29 P30 P31 P32 P33 P34 P35 ...
Embodiment 3
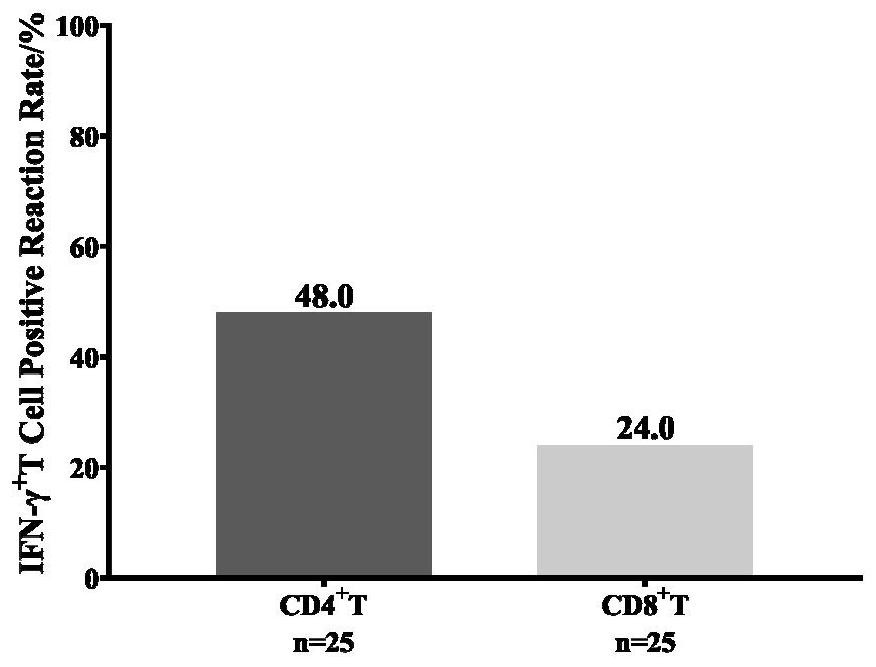
[0042] Example 3 Detection of the proportion of T cells secreting the cytokine IFN-γ
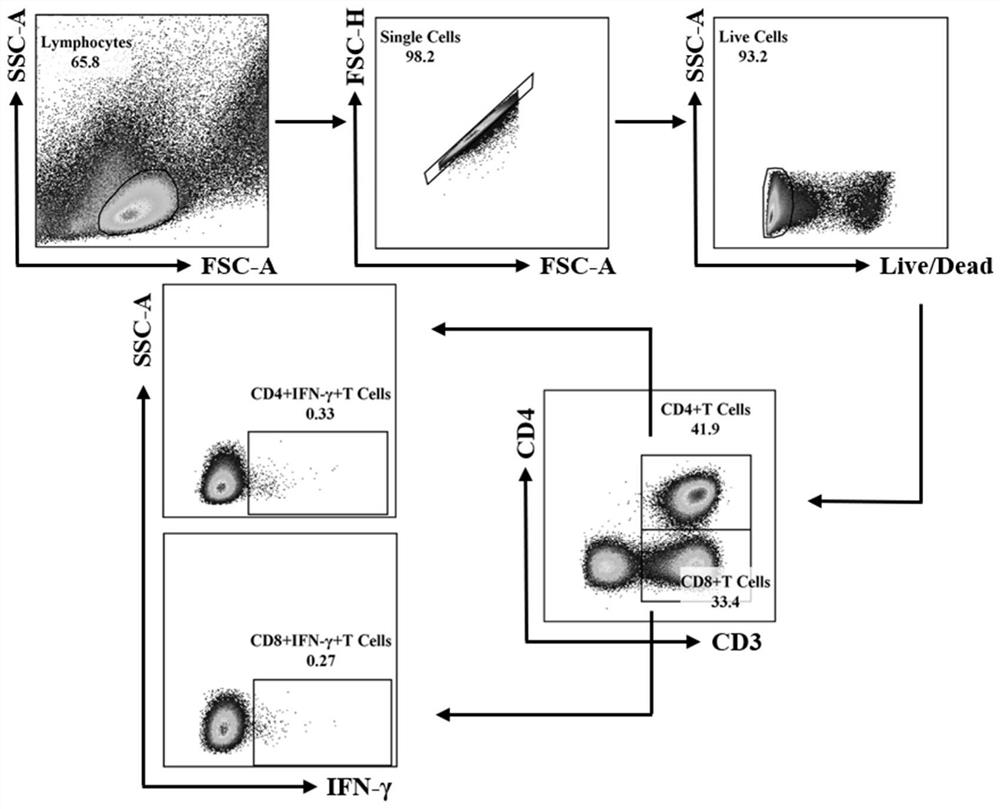
[0043] 1×10 6 Each PBMC was incubated with the peptide group to be tested (the final concentration of each peptide was 2.5 μg / mL) or an equal volume of culture medium (non-stimulation group) for 20 hours (37°C, 5% CO 2 ), Golgi blocker was added 5 hours before the end of incubation. After the end of the culture, the dead cells, cell surface markers (CD3, CD4, CD69) and intracellular cytokines (IFN-γ) were stained according to the reagent instructions, and detected by flow cytometry (BD FACSCantoTMⅡ), and 15- 200,000 lymphocytes were analyzed using the software Flowjo 10, see flow gating strategy figure 1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com